CUTANA™ GST-MeCP2 for meCUT&RUN
{"url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-gst-mecp2-for-me-cut-run","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"options":[{"id":1278,"type":"Configurable_PickList_Set","display_name":"Select Pack Size:","required":true,"condition":true,"state":"variant_option","values":[{"label":"24 Reactions","id":170,"data":"24 Reactions","selected":true},{"label":"96 Reactions","id":171,"data":"96 Reactions","selected":false}],"partial":"set-radio"}],"id":1379,"bulk_discount_rates":[],"can_purchase":true,"meta_description":"","category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1379/1267/meCUTRUN_Icon_For_Product_Pages__87123.1745245422.png?c=2","alt":"CUTANA™ GST-MeCP2 for meCUT&RUN"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1379","shipping":{"calculated":true},"num_reviews":0,"weight":"0.01 LBS","custom_fields":[{"id":"1346","name":"Internal Comment","value":"Excess in bottom of Venom"},{"id":"1347","name":"Internal Comment","value":"Bulk in Psylocke"}],"sku":"15-2002-S","description":"<div class=\"product-general-info\">\n <ul class=\"product-general-info__list-left\">\n <li class=\"product-general-info__list-item\">\n <strong>Type: </strong>Reader Protein\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Mol Wgt: </strong>39.1 kDa\n </li>\n </ul>\n <ul class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <strong>Host: </strong><em>E. coli</em>\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Epitope Tag: </strong>GST, 6xHis\n </li>\n </ul>\n</div>\n<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\" style=\"display: block\">\n <p>\n CUTANA™ GST-MeCP2 for meCUT&RUN is a GST-tagged MeCP2 methyl binding domain that is validated to enrich\n regions of chromatin with symmetrically methylated CpGs in CUTANA™ meCUT&RUN, a modified CUT&RUN workflow\n that enables streamlined, high-resolution mapping of DNA methylation. meCUT&RUN can be followed by a\n traditional library prep method, such as the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher <a\n href=\"/products/epigenetics-kits-and-reagents/cutana-cut-run-library-prep-kit\" target=\"_blank\">14-1001/14-1002</a>),\n to provide ~150 bp resolution profiles of DNA methylation enrichment, or by a cytosine conversion strategy\n such as Enzymatic Methyl-seq (NEB<sup>®</sup> EM-seq™, preferred) or bisulfite sequencing to provide base-pair\n resolution of 5-methylcytosine (5mC).\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\" style=\"display: block\">\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/proteins/15-2002-dna-fragment-size-distribution-analysis.jpeg\" target=\"blank\"\n class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"15-2002-dna-fragment-size-distribution-analysis\"\n src=\"/content/images/products/proteins/15-2002-dna-fragment-size-distribution-analysis.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 1: meCUT&RUN DNA fragment size distribution\n analysis </strong><br />\n meCUT&RUN was performed as described in <strong>Figure 3</strong>. Library DNA was analyzed by\n Agilent TapeStation<sup>®</sup>.\n This analysis confirmed that mononucleosomes were predominantly enriched in meCUT&RUN (~300 bp peaks\n represent 150 bp nucleosomes + sequencing adapters).\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/proteins/15-2002-gene-browser-tracks.jpeg\" target=\"blank\"\n class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"15-2002-gene-browser-tracks\"\n src=\"/content/images/products/proteins/15-2002-gene-browser-tracks.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 2: Gene browser tracks</strong><br />\n meCUT&RUN was performed as described in <strong>Figure 3</strong>. A 30 kb window at the AJM1 gene\n is shown for\n anti-GST antibody and meCUT&RUN. Tracks are also shown with representative data for meCUT&RUN\n followed by EM-seq (meCUT&RUN-EM) and whole genome EM-seq (WGEM), using the New England Biolabs\n NEBNext<sup>®</sup> Enzymatic Methyl-seq v2 Kit (NEB E8015). The meCUT&RUN kit produced the expected genomic\n distribution, showing enrichment of methylated DNA that approximates the methylated CpG pattern\n observed in WGEM. Images were generated using the Integrative Genomics Viewer (IGV, Broad\n Institute).\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/mecut-run-methods.png?t=1745245682\" target=\"blank\"\n class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"meCut&RUN-methods\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/mecut-run-methods.png?t=1745245682\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 3: meCUT&RUN methods</strong><br />\n meCUT&RUN was performed starting with 500k K562 cells with either 2.5 µL of 20X GST-MeCP2 added as\n the primary binding reagent or 0.5 µg of a secondary antibody-only control (anti-GST antibody,\n EpiCypher <a href=\"/products/epigenetics-kits-and-reagents/cutana-anti-gst-tag-antibody\" target=\"_blank\">13-0073</a>) to determine background cleavage. Library preparation was performed using 5 ng of\n meCUT&RUN-enriched DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN\n Library Prep Kit (EpiCypher <a href=\"/products/epigenetics-kits-and-reagents/cutana-cut-run-library-prep-kit\" target=\"_blank\">14-1001/14-1002</a>). Libraries were run on an Illumina NextSeq2000 with\n paired-end sequencing (2x50 bp). Sample sequencing depth was 7.7 million reads (anti-GST) and 8.4\n million reads (GST-MeCP2). Data were aligned to the hg38 genome using Bowtie2. Data were filtered to\n remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img loading=\"lazy\" alt=\"15-2002-dna-fragment-size-distribution-analysis\"\n src=\"/content/images/products/proteins/15-2002-dna-fragment-size-distribution-analysis.jpeg\" width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\" role=\"button\" />\n <img loading=\"lazy\" alt=\"15-2002-gene-browser-tracks\"\n src=\"/content/images/products/proteins/15-2002-gene-browser-tracks.jpeg\" width=\"200\"\n class=\"image-picker__side-image\" role=\"button\" />\n <img loading=\"lazy\" alt=\"meCUT&RUN-methods\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/mecut-run-methods.png?t=1745245682\" width=\"200\"\n class=\"image-picker__side-image\" role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Recommended Accessory Products</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <table class=\"epicypher-table\">\n <tr>\n <th>ITEM</th>\n <th>CAT. NO.</th>\n </tr>\n\n <tr>\n <td>CUTANA™ Anti-GST Tag Antibody</td>\n <td>\n <a\n href=\"/products/epigenetics-kits-and-reagents/cutana-anti-gst-tag-antibody\" target=\"_blank\">13-0073</a>\n </td>\n </tr>\n\n <tr>\n <td>CUTANA™ ChIC/CUT&RUN Kit</td>\n <td>\n <a href=\"/products/epigenetics-kits-and-reagents/cutana-chic-cut-run-kit\" target=\"_blank\">14-1048/14-1048-24</a>\n </td>\n </tr>\n\n <tr>\n <td>CUTANA™ CUT&RUN Library Prep Kit</td>\n <td>\n <a href=\"/products/epigenetics-kits-and-reagents/cutana-cut-run-library-prep-kit\" target=\"_blank\">14-1001/14-1002</a>\n </td>\n </tr>\n\n </table>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Stable for 12 months at -20°C from date of receipt. The protein is not subject to freeze/thaw under these conditions.\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Formulation</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 25 mM HEPES-NaOH pH 6.5, 200 mM NaCl, 1 mM DTT, 50% glycerol, 2 mM MgCl<sub>2</sub>\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\" style=\"outline: none;\">Application Notes</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <p>\n To perform meCUT&RUN with this reagent and a CUTANA CUT&RUN Kit (EpiCypher <a href=\"/products/epigenetics-kits-and-reagents/cutana-chic-cut-run-kit\" target=\"_blank\">14-1048</a>), please follow the <a href=\"https://www.epicypher.com/content/documents/manuals/me-cut-and-run-kit-manual.pdf\" target=\"_blank\">meCUT&RUN Kit Manual</a>. If using homemade buffers, adjust the <a href=\"https://www.epicypher.com/resources/protocols/cutana-pag-mnase-resources\" target=\"_blank\">EpiCypher CUT&RUN Protocol</a> as follows:\n </p>\n <ul style=\"margin-bottom: 0;\"><strong>Day 1</strong>:\n <li>Prepare Pre-Wash Buffer with 200 mM NaCl*. This is used to prepare Wash Buffer, Digitonin Buffer, and Antibody Buffer.</li>\n <ul style=\"margin-bottom: 0;\">\n <li><sup>*</sup><em>NOTE:</em> Standard CUT&RUN uses 150 nM NaCl in these buffers; however, excess salt is recommended to improve MeCP2 specificity for methylated DNA.</li>\n </ul>\n <li>In place of a primary antibody, add 2.5 µL of GST-MeCP2 to each reaction, and incubate on a nutator at 4˚C overnight.</li>\n </ul>\n <ul style=\"margin-bottom: 0;\"><strong>Day 2</strong>:\n <li>On Day 2, discard the GST-MeCP2 supernatant and resuspend each reaction in 50 µL cold Digitonin Buffer.</li>\n <li>Add 1.0 µL anti-GST Tag Antibody (EpiCypher <a href=\"/products/epigenetics-kits-and-reagents/cutana-anti-gst-tag-antibody\" target=\"_blank\">13-0073</a>)* to each reaction and incubate on a nutator at room temperature for 30 minutes.</li>\n <ul style=\"margin-bottom: 0;\"> \n <li><sup>*</sup><em>NOTE:</em> Secondary-only (anti-GST Tag) reactions are recommended as a negative control.</li>\n </ul>\n <li>At the end of the incubation, discard the supernatant and wash two times with 200 µL cold Digitonin Buffer.</li>\n <li>Resuspend in 50 µL per reaction cold Digitonin Buffer and proceed to pAG-MNase (EpiCypher <a href=\"/products/epigenetics-reagents-and-assays/cutana-pag-mnase-for-chic-cut-and-run-workflows\" target=\"_blank\">15-1016/15-1116</a>) addition per the standard protocol.</li>\n </ul>\n <ul><strong>Sequencing Libraries can be prepared using two methods</strong>:\n <li>Option 1: Traditional library prep, which provides a CUT&RUN-like view of genomic regions with high DNA methylation. The CUTANA CUT&RUN Library Prep Kits (EpiCypher <a href=\"/products/epigenetics-kits-and-reagents/cutana-cut-run-library-prep-kit\" target=\"_blank\">14-1001/14-1002</a>) are compatible with this approach.</li>\n <li>Option 2: A cytosine conversion strategy (e.g., New England Biolabs NEBNext<sup>®</sup> Enzymatic Methyl-seq v2 Kit, E8015), which provides base-pair resolution of 5mC enrichment. Prior to EM-seq conversion, add CUTANA™ Fragmented Controls for DNA Methylation Sequencing (EpiCypher <a href=\"/products/epigenetics-kits-and-reagents/cutana-fragmented-controls-dna-methylation-sequencing\" target=\"_blank\">14-1804</a>) to each reaction as outlined in the 14-1804 Technical Data Sheet.</li>\n </ul>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Documents & Resources</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-documents\">\n <a href=\"/content/documents/tds/15-2002-S.pdf\" target=\"blank\" class=\"product-documents__link\">\n <svg version=\"1.1\" id=\"Layer_1\" xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\" x=\"0px\" y=\"0px\" viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\" xml:space=\"preserve\" class=\"product-documents__icon\">\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect x=\"20\" y=\"112\" class=\"product-documents__svg-background\" width=\"146\" height=\"76\" />\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path class=\"product-documents__svg-pdf\" d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Technical Datasheet</span>\n </a>\n </div>\n <div class=\"product-documents\"></div>\n <a href=\"https://www.epicypher.com/content/documents/manuals/me-cut-and-run-kit-manual.pdf\" target=\"blank\"\n class=\"product-documents__link\">\n <svg version=\"1.1\" id=\"Layer_1\" xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\" x=\"0px\" y=\"0px\" viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\" xml:space=\"preserve\" class=\"product-documents__icon\">\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect x=\"20\" y=\"112\" class=\"product-documents__svg-background\" width=\"146\" height=\"76\" />\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path class=\"product-documents__svg-pdf\" d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">meCUT&RUN Manual</span>\n </a>\n </div>\n </div>\n </div>\n </div>\n</div>\n</div>\n\n<style>\n\n .form-field-title .required-text {\n display: none !important;\n }\n\n .form-label {\n padding-left: 2rem;\n }\n\n .form-label-text {\n margin: 0;\n margin-left: 0 !important;\n }\n\n /* //////////// */\n\n /* BIOZ */\n td table {\n margin: 0;\n padding: 6px !important;\n }\n\n span.bioz-w-add-underline {\n font-size: 15px;\n }\n\n /* /////////// */\n\n .form-field-title .required-text {\n display: none !important;\n }\n\n .form-label {\n padding-left: 2rem;\n }\n\n .form-label-text {\n margin: 0;\n margin-left: 0 !important;\n }\n\n .bioz-w-header td {\n padding: 0;\n }\n\n .epicypher-table table {\n margin-right: auto;\n margin-left: auto;\n width: 100%;\n border-top: 1px solid lightgray;\n }\n\n .epicypher-table th {\n color: #3b3a48;\n }\n\n .epicypher-table td {\n color: #3b3a48;\n font-size: 0.9rem;\n }\n\n .epicypher-table td a {\n text-decoration: underline;\n }\n\n .epicypher-table td,\n th {\n border: 0.5px solid lightgray;\n text-align: left;\n padding: 8px;\n /* white-space: nowrap; */\n }\n\n .epicypher-table tr:nth-child(even) {\n background-color: #f9f9f9;\n }\n\n .epicypher-table td:first-child {\n border-left: 1px solid lightgray;\n }\n\n /* tr:hover {\n background-color: #ffff99;\n } */\n\n /* Tablet */\n\n @media only screen and (max-width: 760px),\n (min-device-width: 760px) and (max-device-width: 1024px) {\n\n /* Force table to not be like tables anymore */\n .epicypher-table table,\n thead,\n tbody,\n th,\n td,\n tr {\n display: block;\n }\n\n .epicypher-table th {\n display: none !important;\n }\n\n /* BIOZ */\n td.bioz-w-tooltipx::before {\n display: none;\n }\n\n /* .epicypher-table table,\n thead,\n tbody,\n th,\n td,\n tr {\n display: inline-table !important;\n } */\n\n /* Hide table headers (but not display: none;, for accessibility) */\n .epicypher-table thead tr {\n position: absolute;\n top: -9999px;\n left: -9999px;\n }\n\n .epicypher-table tr {\n border: 1px solid #ccc;\n }\n\n .epicypher-table td {\n /* Behave like a \"row\" */\n /* border: none; */\n border-bottom: 1px solid #eee;\n position: relative;\n padding-left: 30%;\n white-space: inherit;\n }\n\n .epicypher-table td:before {\n /* Now like a table header */\n position: absolute;\n /* Top/left values mimic padding */\n top: 6px;\n left: 6px;\n width: 45%;\n padding-right: 10px;\n white-space: nowrap;\n font-weight: 700;\n }\n\n /*\n Label the data\n */\n .epicypher-table td:nth-of-type(1):before {\n content: 'Item';\n }\n\n td:nth-of-type(2):before {\n content: 'Cat. No.';\n }\n }\n\n /* @media screen and (max-width: 767px) {\n .custom-title-description {\n font-size: 19px !important;\n }\n .section-title {\n font-size: 19px !important;\n padding-left: 1rem;\n }\n } */\n\n @media screen and (min-width: 768px) {\n\n .products-featured .container,\n .products-featured .product-tabs,\n .products-related .container,\n .products-related .product-tabs {\n padding-left: 30px !important;\n padding-right: 50px;\n max-width: 1170px !important;\n }\n }\n\n @media screen and (max-width: 767px) {\n .custom-title-description {\n font-size: 19px !important;\n }\n\n .section-title {\n font-size: 19px !important;\n padding-left: 1rem;\n }\n }\n</style>","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$245.00","value":245,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"CUTANA™ GST-MeCP2 for meCUT&RUN | EpiCypher","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"shipping_messages":[],"rating":0,"meta_keywords":"mecp2, DNA methylation, meCUT&RUN, CUT&RUN, CUT and RUN, genomic mapping, ChIP-Seq","show_quantity_input":1,"title":"CUTANA™ GST-MeCP2 for meCUT&RUN","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1379/1267/meCUTRUN_Icon_For_Product_Pages__87123.1745245422.png?c=2","alt":"CUTANA™ GST-MeCP2 for meCUT&RUN"}]}
- Type: Reader Protein
- Mol Wgt: 39.1 kDa
- Host: E. coli
- Epitope Tag: GST, 6xHis
Description
CUTANA™ GST-MeCP2 for meCUT&RUN is a GST-tagged MeCP2 methyl binding domain that is validated to enrich regions of chromatin with symmetrically methylated CpGs in CUTANA™ meCUT&RUN, a modified CUT&RUN workflow that enables streamlined, high-resolution mapping of DNA methylation. meCUT&RUN can be followed by a traditional library prep method, such as the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002), to provide ~150 bp resolution profiles of DNA methylation enrichment, or by a cytosine conversion strategy such as Enzymatic Methyl-seq (NEB® EM-seq™, preferred) or bisulfite sequencing to provide base-pair resolution of 5-methylcytosine (5mC).
Validation Data
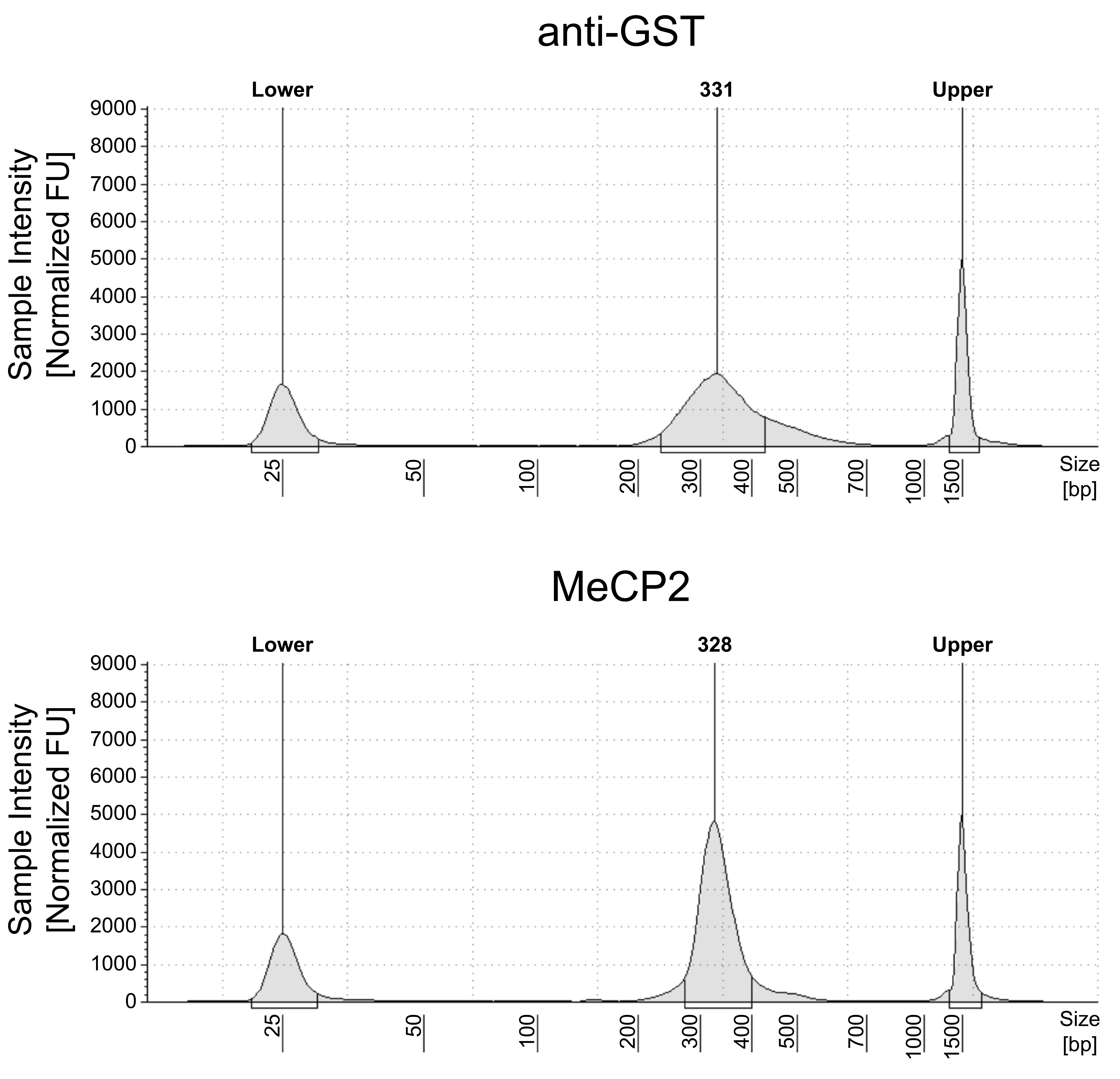
Figure 1: meCUT&RUN DNA fragment size distribution
analysis
meCUT&RUN was performed as described in Figure 3. Library DNA was analyzed by
Agilent TapeStation®.
This analysis confirmed that mononucleosomes were predominantly enriched in meCUT&RUN (~300 bp peaks
represent 150 bp nucleosomes + sequencing adapters).
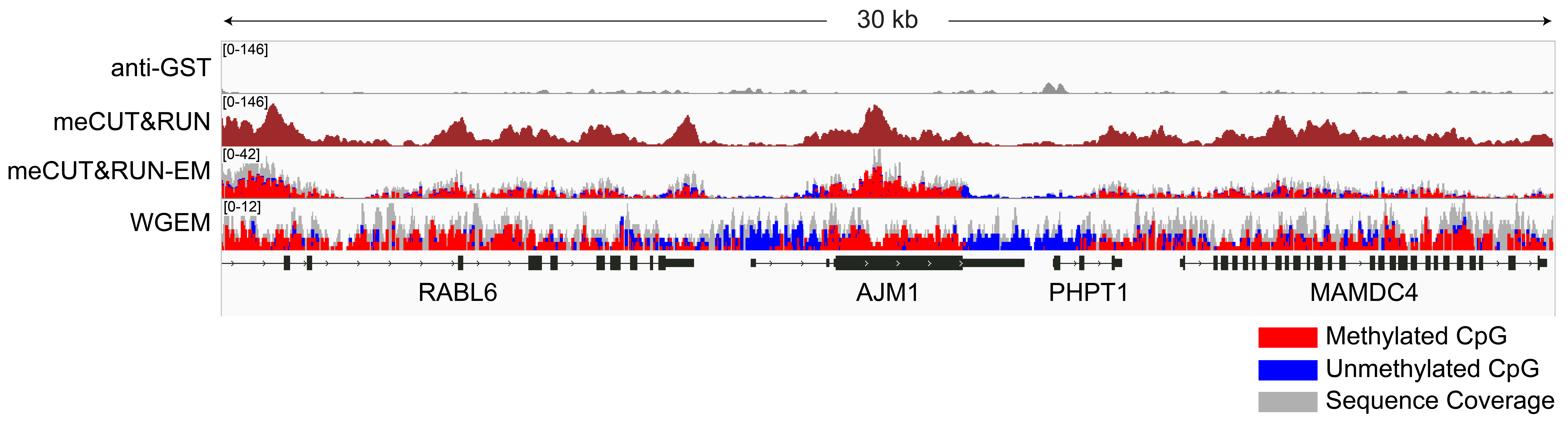
Figure 2: Gene browser tracks
meCUT&RUN was performed as described in Figure 3. A 30 kb window at the AJM1 gene
is shown for
anti-GST antibody and meCUT&RUN. Tracks are also shown with representative data for meCUT&RUN
followed by EM-seq (meCUT&RUN-EM) and whole genome EM-seq (WGEM), using the New England Biolabs
NEBNext® Enzymatic Methyl-seq v2 Kit (NEB E8015). The meCUT&RUN kit produced the expected genomic
distribution, showing enrichment of methylated DNA that approximates the methylated CpG pattern
observed in WGEM. Images were generated using the Integrative Genomics Viewer (IGV, Broad
Institute).
Figure 3: meCUT&RUN methods
meCUT&RUN was performed starting with 500k K562 cells with either 2.5 µL of 20X GST-MeCP2 added as
the primary binding reagent or 0.5 µg of a secondary antibody-only control (anti-GST antibody,
EpiCypher 13-0073) to determine background cleavage. Library preparation was performed using 5 ng of
meCUT&RUN-enriched DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN
Library Prep Kit (EpiCypher 14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with
paired-end sequencing (2x50 bp). Sample sequencing depth was 7.7 million reads (anti-GST) and 8.4
million reads (GST-MeCP2). Data were aligned to the hg38 genome using Bowtie2. Data were filtered to
remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
Recommended Accessory Products
| ITEM | CAT. NO. |
|---|---|
| CUTANA™ Anti-GST Tag Antibody | 13-0073 |
| CUTANA™ ChIC/CUT&RUN Kit | 14-1048/14-1048-24 |
| CUTANA™ CUT&RUN Library Prep Kit | 14-1001/14-1002 |
Technical Information
Application Notes
To perform meCUT&RUN with this reagent and a CUTANA CUT&RUN Kit (EpiCypher 14-1048), please follow the meCUT&RUN Kit Manual. If using homemade buffers, adjust the EpiCypher CUT&RUN Protocol as follows:
- Day 1:
- Prepare Pre-Wash Buffer with 200 mM NaCl*. This is used to prepare Wash Buffer, Digitonin Buffer, and Antibody Buffer.
- *NOTE: Standard CUT&RUN uses 150 nM NaCl in these buffers; however, excess salt is recommended to improve MeCP2 specificity for methylated DNA.
- In place of a primary antibody, add 2.5 µL of GST-MeCP2 to each reaction, and incubate on a nutator at 4˚C overnight.
- Day 2:
- On Day 2, discard the GST-MeCP2 supernatant and resuspend each reaction in 50 µL cold Digitonin Buffer.
- Add 1.0 µL anti-GST Tag Antibody (EpiCypher 13-0073)* to each reaction and incubate on a nutator at room temperature for 30 minutes.
- *NOTE: Secondary-only (anti-GST Tag) reactions are recommended as a negative control.
- At the end of the incubation, discard the supernatant and wash two times with 200 µL cold Digitonin Buffer.
- Resuspend in 50 µL per reaction cold Digitonin Buffer and proceed to pAG-MNase (EpiCypher 15-1016/15-1116) addition per the standard protocol.
- Sequencing Libraries can be prepared using two methods:
- Option 1: Traditional library prep, which provides a CUT&RUN-like view of genomic regions with high DNA methylation. The CUTANA CUT&RUN Library Prep Kits (EpiCypher 14-1001/14-1002) are compatible with this approach.
- Option 2: A cytosine conversion strategy (e.g., New England Biolabs NEBNext® Enzymatic Methyl-seq v2 Kit, E8015), which provides base-pair resolution of 5mC enrichment. Prior to EM-seq conversion, add CUTANA™ Fragmented Controls for DNA Methylation Sequencing (EpiCypher 14-1804) to each reaction as outlined in the 14-1804 Technical Data Sheet.