CUTANA™ Anti-GST Tag Antibody
{"url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-anti-gst-tag-antibody","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"id":1377,"bulk_discount_rates":[],"can_purchase":true,"meta_description":"","category":["Antibodies","Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays","Antibodies/CUTANA™ CUT&RUN Antibodies"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1377/1268/meCUTRUN_Icon_For_Product_Pages__47683.1745214123.png?c=2","alt":"CUTANA™ Anti-GST Tag Antibody"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1377","shipping":{"calculated":true},"num_reviews":0,"weight":"0.00 LBS","custom_fields":[{"id":"1348","name":"Pack Size","value":"100 µg"}],"sku":"13-0073","description":"<div class=\"product-general-info\">\n <ul class=\"product-general-info__list-left\">\n <li class=\"product-general-info__list-item\">\n <strong>Type: </strong>Polyclonal\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Host: </strong>Rabbit\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Applications: </strong>CUT&RUN, WB\n </li>\n </ul>\n <ul class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <strong>Reactivity: </strong>GST Tag\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Format: </strong>Purified\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Target Size: </strong>N/A\n </li>\n </ul>\n <!-- <ul style=\"padding: 0\" class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <a href=\"#bioz\">\n <div id=\"w-s-3835-13-2010\" style=\"\n width: 240px;\n height: 58px;\n position: relative;\n overflow-y: hidden;\n \"></div>\n <div id=\"bioz-w-pb-13-2010-div\" style=\"width: 240px\">\n <a id=\"bioz-w-pb-13-2010\" style=\"font-size: 12px; color: transparent\" href=\"https://www.bioz.com/\"\n target=\"_blank\">\n <img src=\"https://cdn.bioz.com/assets/favicon.png\" style=\"\n width: 11px;\n height: 11px;\n vertical-align: baseline;\n padding-bottom: 0px;\n margin-left: 0px;\n margin-bottom: 0px;\n float: none;\n display: none;\n \" />\n Powered by Bioz\n </a>\n </div>\n </a>\n </li>\n </ul> -->\n</div>\n<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <p>\n Unlock the full potential of Reader CUT&RUN with our high-affinity CUTANA™ Anti-GST Tag Antibody,\n specifically developed to support chromatin profiling applications wherein a GST-tagged reader protein\n replaces the traditional primary antibody in directing the selective cleavage and release of target chromatin\n fragments by pAG-MNase [1]. For example, this antibody is used in CUTANA™ meCUT&RUN, a modified CUT&RUN\n workflow in which a GST-tagged MeCP2 methyl binding domain is used to enrich regions of chromatin with\n symmetrically methylated CpGs, enabling streamlined, high-resolution mapping of DNA methylation.\n </p>\n <p>\n Engineered for superior performance in low-background, high-resolution CUT&RUN assays, this antibody offers\n exceptional sensitivity and specificity for detecting GST-fusion proteins bound to chromatin. Whether\n studying histone modifications, DNA methylation, or other epigenetic readers, this antibody is the ideal\n tool for enabling precise, reproducible mapping using GST-tagged fusion constructs.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/antibodies/13-0073-dna-fragment-size-distribution-analysis.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"13-0073-dna-fragment-size-distribution-analysis\"\n src=\"/content/images/products/antibodies/13-0073-dna-fragment-size-distribution-analysis.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 1: meCUT&RUN DNA fragment size distribution\n analysis</strong><br />\n\n meCUT&RUN was performed as described in <strong>Figure 4</strong>. Library DNA was analyzed by\n Agilent TapeStation<sup>®</sup>.\n This analysis confirmed that mononucleosomes were predominantly enriched in meCUT&RUN (~300 bp peaks\n represent 150 bp nucleosomes + sequencing adapters).\n\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/antibodies/13-0073-gene-browser-tracks.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"13-0073-gene-browser-tracks\"\n src=\"/content/images/products/antibodies/13-0073-gene-browser-tracks.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 2: Gene browser tracks</strong><br />\n meCUT&RUN was performed as described in <strong>Figure 4</strong>. A 30 kb window at the AJM1 gene\n is shown for\n anti-GST antibody and meCUT&RUN. Tracks are also shown with representative data for meCUT&RUN\n followed by EM-seq (meCUT&RUN-EM) and whole genome EM-seq (WGEM), using the New England Biolabs\n NEBNext<sup>®</sup> Enzymatic Methyl-seq v2 Kit (NEB E8015). The meCUT&RUN kit produced the expected genomic\n distribution, showing enrichment of methylated DNA that approximates the methylated CpG pattern\n observed in WGEM. Images were generated using the Integrative Genomics Viewer (IGV, Broad\n Institute).\n </span>\n </p>\n </div>\n\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/antibodies/13-0073-western-blot.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"13-0073-western-blot\"\n src=\"/content/images/products/antibodies/13-0073-western-blot.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 3: Western blot data</strong><br />\n Western analysis of GST Tag in 0.1, 0.3, or 1.0 µg of HEK293 cell lysate expressing a GST-tag fusion\n protein. Lysate was resolved via SDS-PAGE and detected with a 1:7,500 dilution of EpiCypher anti-GST\n Tag antibody.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/mecut-run-methods.png?t=1745245682\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"13-2013_western_blot\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/mecut-run-methods.png?t=1745245682\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 4: meCUT&RUN methods\n </strong><br />\n meCUT&RUN was performed starting with 500k K562 cells with either 2.5 µL of 20X GST-MeCP2 (EpiCypher\n <a href=\"/products/epigenetics-kits-and-reagents/cutana-gst-mecp2-for-me-cut-run\"\n target=\"_blank\">15-2002</a>) added as the primary binding reagent or 0.5 µg of a secondary\n antibody-only control\n (anti-GST antibody) to determine background cleavage. Library preparation was performed using 5 ng of\n meCUT&RUN-enriched DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN\n Library Prep Kit (EpiCypher <a\n href=\"/products/epigenetics-kits-and-reagents/cutana-cut-run-library-prep-kit\"\n target=\"_blank\">14-1001/14-1002</a>). Libraries were run on an Illumina NextSeq2000 with\n paired-end sequencing (2x50 bp). Sample sequencing depth was 7.7 million reads (anti-GST) and 8.4\n million reads (GST-MeCP2). Data were aligned to the hg38 genome using Bowtie2. Data were filtered to\n remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.\n </span>\n </p>\n </div>\n </div> \n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img loading=\"lazy\" alt=\"13-0073-dna-fragment-size-distribution-analysis\"\n src=\"/content/images/products/antibodies/13-0073-dna-fragment-size-distribution-analysis.jpeg\" width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\" role=\"button\" />\n <img loading=\"lazy\" alt=\"13-0073-gene-browser-tracks\"\n src=\"/content/images/products/antibodies/13-0073-gene-browser-tracks.jpeg\" class=\"image-picker__side-image\"\n role=\"button\" />\n <img loading=\"lazy\" alt=\"13-0073-western-blot\"\n src=\"/content/images/products/antibodies/13-0073-western-blot.jpeg\" class=\"image-picker__side-image\"\n role=\"button\" />\n <img loading=\"lazy\" alt=\"meCUT&RUN-methods\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/mecut-run-methods.png?t=1745245682\" class=\"image-picker__side-image\"\n role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Immunogen</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Glutathione-S-Transferase (GST)\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Stable for 1 year at 4°C from date of receipt\n\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Formulation</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Antigen affinity-purified antibody in phosphate buffered saline (PBS), 0.09% sodium azide\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Recommended Dilution</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>CUT&RUN:</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 0.5 µg per reaction\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Western Blot (WB):</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 1:500 – 1:15,000\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Recommended Accessory Products</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <table class=\"epicypher-table\">\n <tr>\n <th>ITEM</th>\n <th>CAT. NO.</th>\n </tr>\n\n <tr>\n <td>GST-MeCP2 for meCUT&RUN</td>\n <td>\n <a\n href=\"/products/epigenetics-kits-and-reagents/cutana-gst-mecp2-for-me-cut-run\" target=\"_blank\">15-2002</a>\n </td>\n </tr>\n\n <tr>\n <td>CUTANA™ ChIC/CUT&RUN Kit</td>\n <td>\n <a href=\"/products/epigenetics-kits-and-reagents/cutana-chic-cut-run-kit\" target=\"_blank\">14-1048/14-1048-24</a>\n </td>\n </tr>\n\n <tr>\n <td>CUTANA™ CUT&RUN Library Prep Kit</td>\n <td>\n <a href=\"/products/epigenetics-kits-and-reagents/cutana-cut-run-library-prep-kit\" target=\"_blank\">14-1001/14-1002</a>\n </td>\n </tr>\n\n </table>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">References</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <strong>Background References:</strong>\n <br />\n [1] Marunde et al. <em>eLife</em> (2024). PMID:\n <a href=\"https://pubmed.ncbi.nlm.nih.gov/38319148/\"\n title=\"Nucleosome conformation dictates the histone code\" target=\"_blank\">\n 38319148</a><br />\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Documents & Resources</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-documents\">\n <a href=\"/content/documents/tds/13-0073.pdf\" target=\"_blank\" class=\"product-documents__link\">\n <svg version=\"1.1\" id=\"Layer_1\" xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\" x=\"0px\" y=\"0px\" viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\" xml:space=\"preserve\" class=\"product-documents__icon\">\n <g>\n <path class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect x=\"20\" y=\"112\" class=\"product-documents__svg-background\" width=\"146\" height=\"76\" />\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Technical Datasheet</span>\n </a>\n </div>\n </div>\n </div>\n </div>\n\n <!-- <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 id=\"bioz\" style=\"padding-bottom: 5px; margin-top: 1rem\" class=\"sub-title1\">\n Product References\n </h3>\n\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <object id=\"wobj-3835-13-2010-q\" type=\"text/html\" data=\"https://www.bioz.com/v_widget_6_0/3835/13-2010/\"\n style=\"width: 100%; height: 193px\"></object>\n <div id=\"bioz-w-pb-3835-13-2010-q-div\" style=\"width: 100%\">\n <a id=\"bioz-w-pb-3835-13-2010-q\" style=\"\n font-size: 12px;\n text-decoration: none;\n color: #4698cb;\n \" href=\"https://www.bioz.com/\" target=\"_blank\"><img src=\"https://cdn.bioz.com/assets/favicon.png\"\n style=\"\n width: 11px;\n height: 11px;\n vertical-align: baseline;\n padding-bottom: 0px;\n margin-left: 0px;\n margin-bottom: 0px;\n float: none;\n \" />\n Powered by Bioz</a>\n <a style=\"\n font-size: 12px;\n text-decoration: none;\n float: right;\n color: transparent;\n \" href=\"https://www.bioz.com/result/13-2010/product/EpiCypher/?cn=13-2010\" target=\"_blank\">\n See more details on Bioz</a>\n </div>\n </div>\n </div>\n </div> -->\n </div>\n</div>\n<!-- <script>\n $(document).ready(function () {\n var widget_micro_obj = new v_widget_obj(\"s\", [1]);\n widget_micro_obj.request_catalog_number_widget_data_internal(\n \"13-2031\",\n \"13-2031\"\n );\n });\n</script> -->\n<style>\n /* .bioz-w-header td {\n padding: 0;\n } */\n\n td table {\n margin: 0;\n padding: 6px !important;\n }\n\n .epicypher-table table {\n margin-right: auto;\n margin-left: auto;\n width: 100%;\n border-top: 1px solid lightgray;\n }\n\n .epicypher-table th {\n color: #3b3a48;\n }\n\n .epicypher-table td {\n color: #3b3a48;\n font-size: 0.9rem;\n }\n\n .epicypher-table td a {\n text-decoration: underline;\n }\n\n .epicypher-table td,\n th {\n border: 0.5px solid lightgray;\n text-align: left;\n padding: 8px;\n /* white-space: nowrap; */\n }\n\n .epicypher-table tr:nth-child(even) {\n background-color: #f9f9f9;\n }\n\n .epicypher-table td:first-child {\n border-left: 1px solid lightgray;\n }\n\n @media (min-width:1024px) {\n .product-general-info .product-general-info__list-item {\n white-space: nowrap;\n }\n }\n\n @media only screen and (max-width: 1024px) {\n\n /* Force table to not be like tables anymore */\n .epicypher-table th {\n display: none !important;\n }\n\n /* Hide table headers (but not display: none;, for accessibility) */\n .epicypher-table thead tr {\n position: absolute;\n top: -9999px;\n left: -9999px;\n }\n\n .epicypher-table tr {\n border: 1px solid #ccc;\n }\n\n .epicypher-table td {\n /* Behave like a \"row\" */\n /* border: none; */\n border-bottom: 1px solid #eee;\n position: relative;\n padding-left: 30%;\n white-space: inherit;\n }\n\n .epicypher-table td:before {\n /* Now like a table header */\n position: absolute;\n /* Top/left values mimic padding */\n top: 6px;\n left: 6px;\n width: 45%;\n padding-right: 10px;\n white-space: nowrap;\n font-weight: 700;\n }\n\n /*\n Label the data\n */\n .epicypher-table td:nth-of-type(1):before {\n content: \"Item\";\n }\n\n td:nth-of-type(2):before {\n content: \"Cat. No.\";\n }\n }\n\n \n</style>","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$250.00","value":250,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"CUTANA™ Anti-GST Tag Antibody","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"options":[],"related_products":[{"id":889,"sku":null,"name":"CUTANA™ CUT&RUN Library Prep Kit","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-cut-run-library-prep-kit","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n \n \n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/889/997/Epicypher_CUTANA_CUTRUN_LibraryPrep_Box__64431.1646406521.png?c=2","alt":"CUTANA™ CUT&RUN Library Prep Kit"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/889/997/Epicypher_CUTANA_CUTRUN_LibraryPrep_Box__64431.1646406521.png?c=2","alt":"CUTANA™ CUT&RUN Library Prep Kit"}],"date_added":"31st Jan 2022","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":983,"name":"Pack Size","value":"48 Reactions"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$1,525.00","value":1525},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=889"},{"id":694,"sku":null,"name":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows","url":"https://www.epicypher.com/products/epigenetics-reagents-and-assays/cutana-pag-mnase-for-chic-cut-and-run-workflows","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n Type: Nuclease\n \n \n Mol Wgt: 43.7 kDa\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/694/689/Screen_Shot_2020-02-12_at_11.01.55_AM__17144.1581530752.png?c=2","alt":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/694/689/Screen_Shot_2020-02-12_at_11.01.55_AM__17144.1581530752.png?c=2","alt":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows"}],"date_added":"12th Aug 2019","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1174,"name":"Internal Comment","value":"Excess in bottom of Venom"},{"id":1175,"name":"Internal Comment","value":"Bulk in Psylocke"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$335.00","value":335},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$335.00","value":335},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$1,295.00","value":1295},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=694"},{"id":758,"sku":"13-0042","name":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag","url":"https://www.epicypher.com/products/nucleosomes/snap-cutana-spike-in-controls/cutana-igg-negative-control-antibody-for-cut-run-and-cut-tag","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes/SNAP-CUTANA™ Spike-in Controls","Antibodies/CUTANA™ CUT&RUN Antibodies","Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n Type: Polyclonal\n \n \n Host: Rabbit\n \n \n Applications: CUT&R","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/758/739/1579731877.1280.1280__61771.1592491196.png?c=2","alt":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/758/739/1579731877.1280.1280__61771.1592491196.png?c=2","alt":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag"}],"date_added":"17th Jun 2020","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1148,"name":"Pack Size","value":"100 µg"},{"id":1149,"name":"Internal Comment","value":"bulk stocks from Thermo in cold room"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=758","price":{"without_tax":{"currency":"USD","formatted":"$105.00","value":105},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=758"}],"shipping_messages":[],"rating":0,"meta_keywords":"anti-GST antibody, CUT&RUN antibody, meCUT&RUN, reader CUT&RUN, GST, GST Tag","show_quantity_input":1,"title":"CUTANA™ Anti-GST Tag Antibody","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1377/1268/meCUTRUN_Icon_For_Product_Pages__47683.1745214123.png?c=2","alt":"CUTANA™ Anti-GST Tag Antibody"}]} Pack Size: 100 µg
- Type: Polyclonal
- Host: Rabbit
- Applications: CUT&RUN, WB
- Reactivity: GST Tag
- Format: Purified
- Target Size: N/A
Description
Unlock the full potential of Reader CUT&RUN with our high-affinity CUTANA™ Anti-GST Tag Antibody, specifically developed to support chromatin profiling applications wherein a GST-tagged reader protein replaces the traditional primary antibody in directing the selective cleavage and release of target chromatin fragments by pAG-MNase [1]. For example, this antibody is used in CUTANA™ meCUT&RUN, a modified CUT&RUN workflow in which a GST-tagged MeCP2 methyl binding domain is used to enrich regions of chromatin with symmetrically methylated CpGs, enabling streamlined, high-resolution mapping of DNA methylation.
Engineered for superior performance in low-background, high-resolution CUT&RUN assays, this antibody offers exceptional sensitivity and specificity for detecting GST-fusion proteins bound to chromatin. Whether studying histone modifications, DNA methylation, or other epigenetic readers, this antibody is the ideal tool for enabling precise, reproducible mapping using GST-tagged fusion constructs.
Validation Data
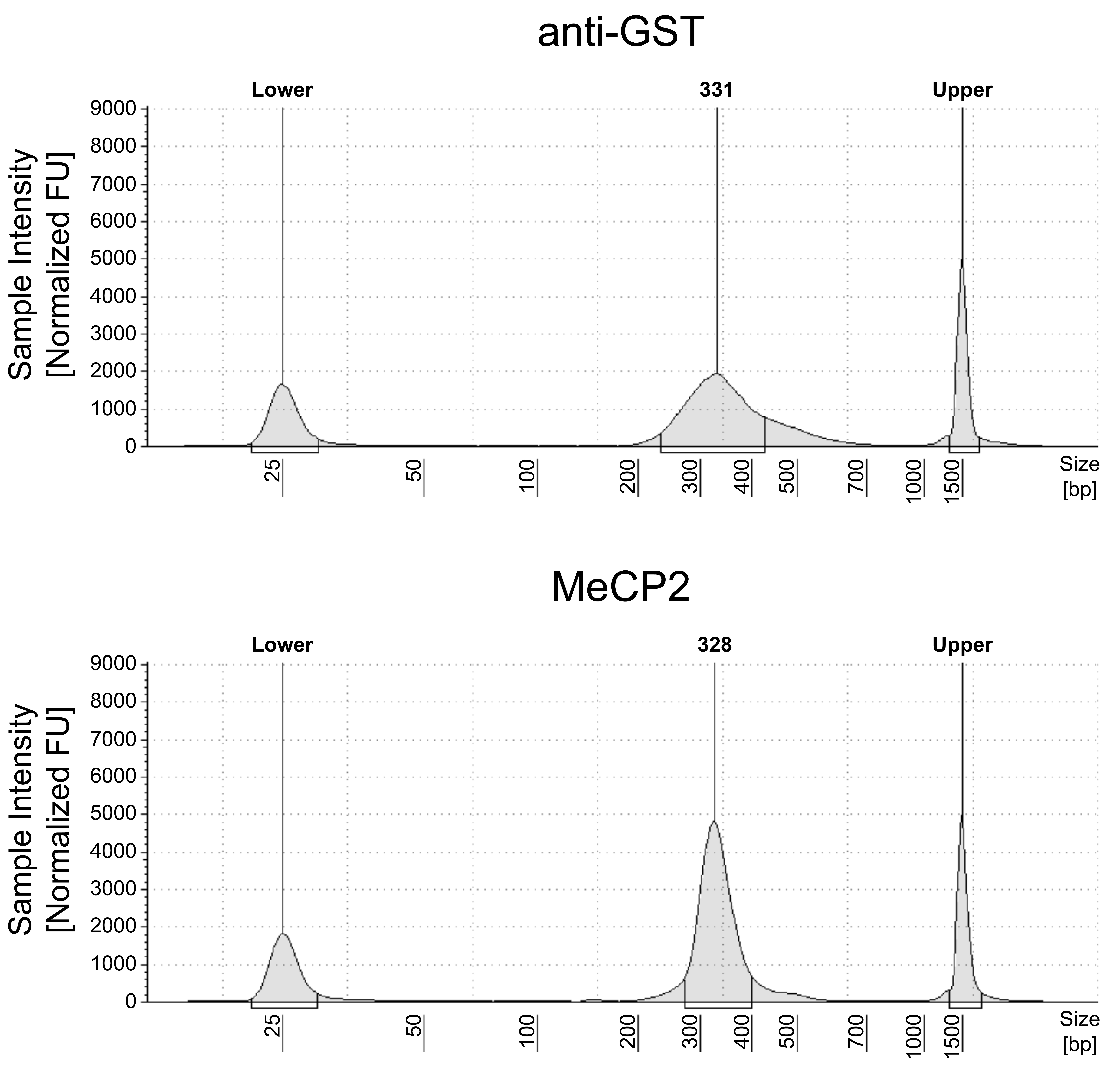
Figure 1: meCUT&RUN DNA fragment size distribution
analysis
meCUT&RUN was performed as described in Figure 4. Library DNA was analyzed by
Agilent TapeStation®.
This analysis confirmed that mononucleosomes were predominantly enriched in meCUT&RUN (~300 bp peaks
represent 150 bp nucleosomes + sequencing adapters).
Figure 2: Gene browser tracks
meCUT&RUN was performed as described in Figure 4. A 30 kb window at the AJM1 gene
is shown for
anti-GST antibody and meCUT&RUN. Tracks are also shown with representative data for meCUT&RUN
followed by EM-seq (meCUT&RUN-EM) and whole genome EM-seq (WGEM), using the New England Biolabs
NEBNext® Enzymatic Methyl-seq v2 Kit (NEB E8015). The meCUT&RUN kit produced the expected genomic
distribution, showing enrichment of methylated DNA that approximates the methylated CpG pattern
observed in WGEM. Images were generated using the Integrative Genomics Viewer (IGV, Broad
Institute).
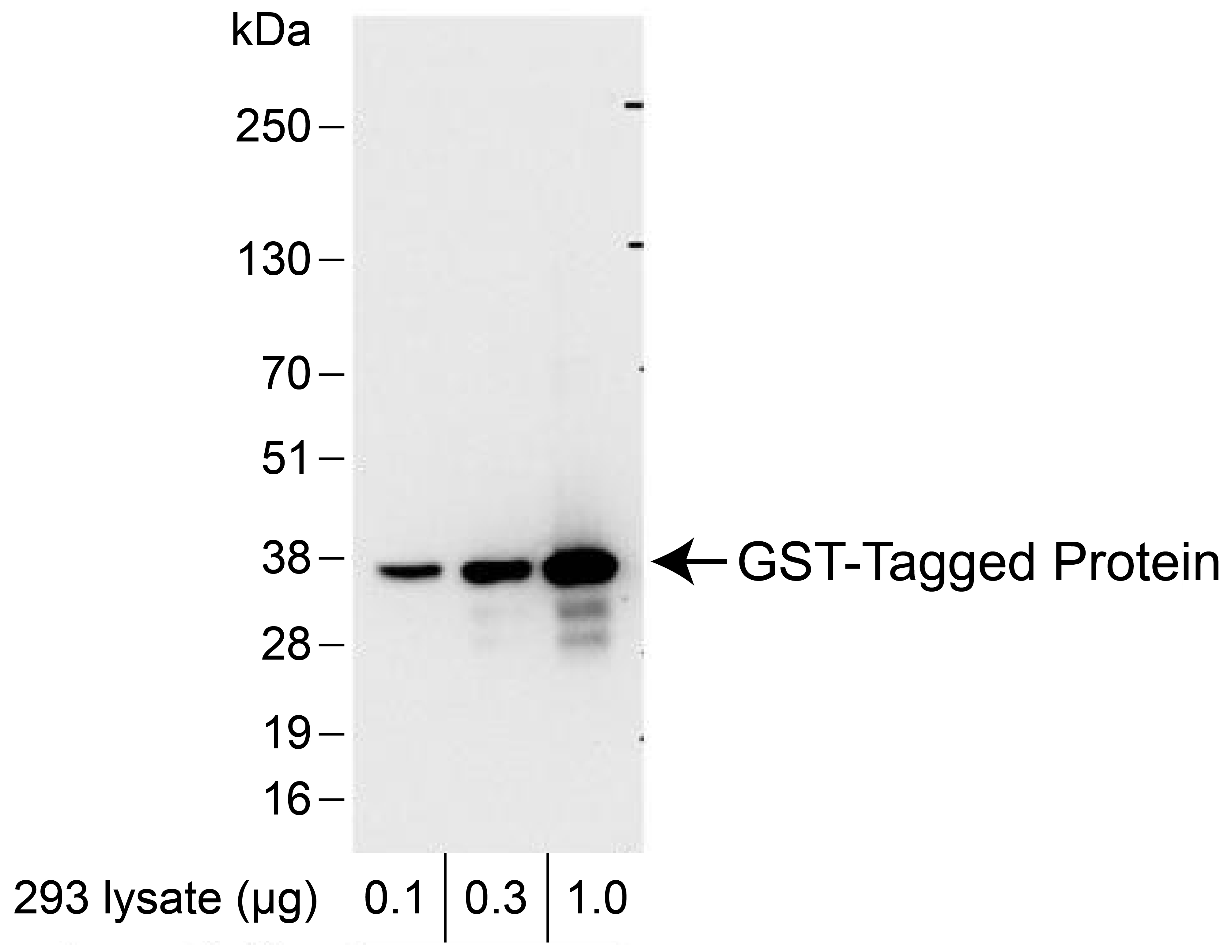
Figure 3: Western blot data
Western analysis of GST Tag in 0.1, 0.3, or 1.0 µg of HEK293 cell lysate expressing a GST-tag fusion
protein. Lysate was resolved via SDS-PAGE and detected with a 1:7,500 dilution of EpiCypher anti-GST
Tag antibody.
Figure 4: meCUT&RUN methods
meCUT&RUN was performed starting with 500k K562 cells with either 2.5 µL of 20X GST-MeCP2 (EpiCypher
15-2002) added as the primary binding reagent or 0.5 µg of a secondary
antibody-only control
(anti-GST antibody) to determine background cleavage. Library preparation was performed using 5 ng of
meCUT&RUN-enriched DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN
Library Prep Kit (EpiCypher 14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with
paired-end sequencing (2x50 bp). Sample sequencing depth was 7.7 million reads (anti-GST) and 8.4
million reads (GST-MeCP2). Data were aligned to the hg38 genome using Bowtie2. Data were filtered to
remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
Technical Information
Recommended Dilution
Recommended Accessory Products
| ITEM | CAT. NO. |
|---|---|
| GST-MeCP2 for meCUT&RUN | 15-2002 |
| CUTANA™ ChIC/CUT&RUN Kit | 14-1048/14-1048-24 |
| CUTANA™ CUT&RUN Library Prep Kit | 14-1001/14-1002 |