CUTANA™ ChIC/CUT&RUN Kit
Description
The CUTANA™ ChIC/CUT&RUN Kit enables streamlined chromatin profiling of histone post-translational modifications (PTMs) and chromatin associated proteins.
Save 10% when you buy with our Library Prep Kit for a streamlined CUT&RUN workflow! Learn more >>
The CUT&RUN Kit Version 5 (v5) now includes an additional control antibody (H3K27me3). Positive (H3K4me3 and H3K27me3) and negative (IgG) control antibodies pair with SNAP-CUTANA™ spike-in controls for assay optimization and continuous assay monitoring (Figure 2). E. coli DNA is included for data normalization. SPRI magnetic beads are used for DNA purification, enabling seamless multi-channel pipetting throughout the workflow to maximize throughput and reproducibility. The kit is compatible with a variety of inputs including cells or nuclei derived from native, cryopreserved, or cross-linked samples. While it is recommended to start with 500,000 cells, comparable data can be generated using as few as 5,000 cells. The inclusion of controls, as well as compatibility with diverse target types, sample inputs, and low cell numbers, make this kit the go-to solution for chromatin mapping experiments. To place bulk orders, contact info@epicypher.com.
Keep learning by checking out our Tech Support Center.
Validation Data
Figure 1: CUT&RUN DNA fragment size distribution
analysis
CUT&RUN was performed as described in
Figure 5. Library DNA was analyzed by
Agilent Tapestation®. This analysis
confirmed that mononucleosomes were predominantly
enriched in CUT&RUN (~300 bp peaks represent 150 bp
nucleosomes + sequencing adapters).
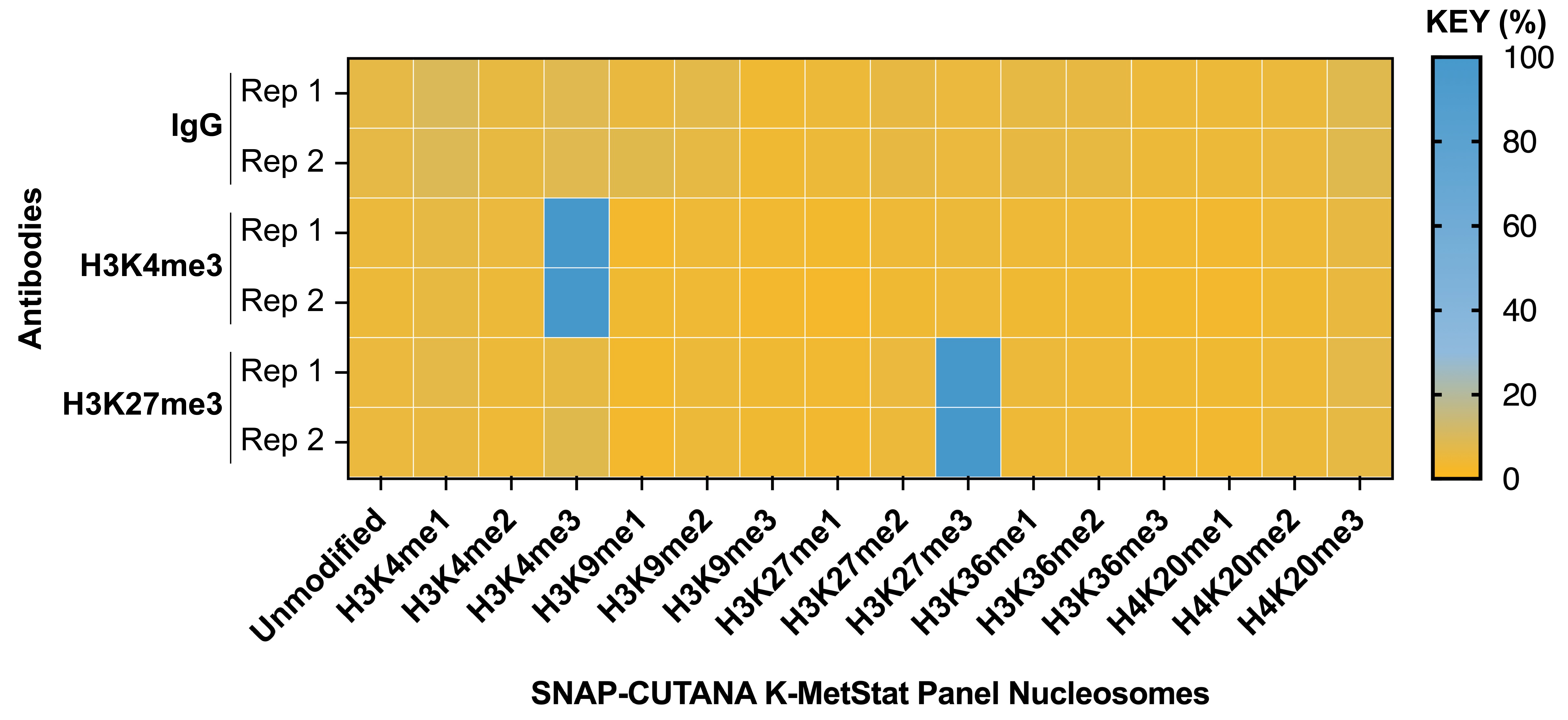
Figure 2: SNAP-CUTANA™ Spike-in
controls
DNA-barcoded designer nucleosomes (dNucs) harboring distinct K-methyl PTMs were spiked into CUT&RUN
reactions prior to antibody addition. Spike-in barcodes were analyzed using the shell script
available on the spike-in product page (EpiCypher 19-1002). Barcodes for IgG (normalized to total
reads), H3K4me3 and H3K27me3 (normalized to on-target) antibodies are shown. The spike-ins confirmed
H3K4me3 and H3K27me3 antibodies specifically recovered the target dNucs, while IgG showed no
preferential enrichment.
Figure 3: CUT&RUN genome-wide heatmaps
CUT&RUN was performed as described in Figure 5. Heatmaps show two replicates
(“Rep”) of IgG,
H3K4me3, and H3K27me3 kit control antibodies in aligned rows ranked by intensity (top to bottom)
relative to the H3K4me3 Rep 1 reaction and colored such that red indicates high localized enrichment
and blue denotes background signal.
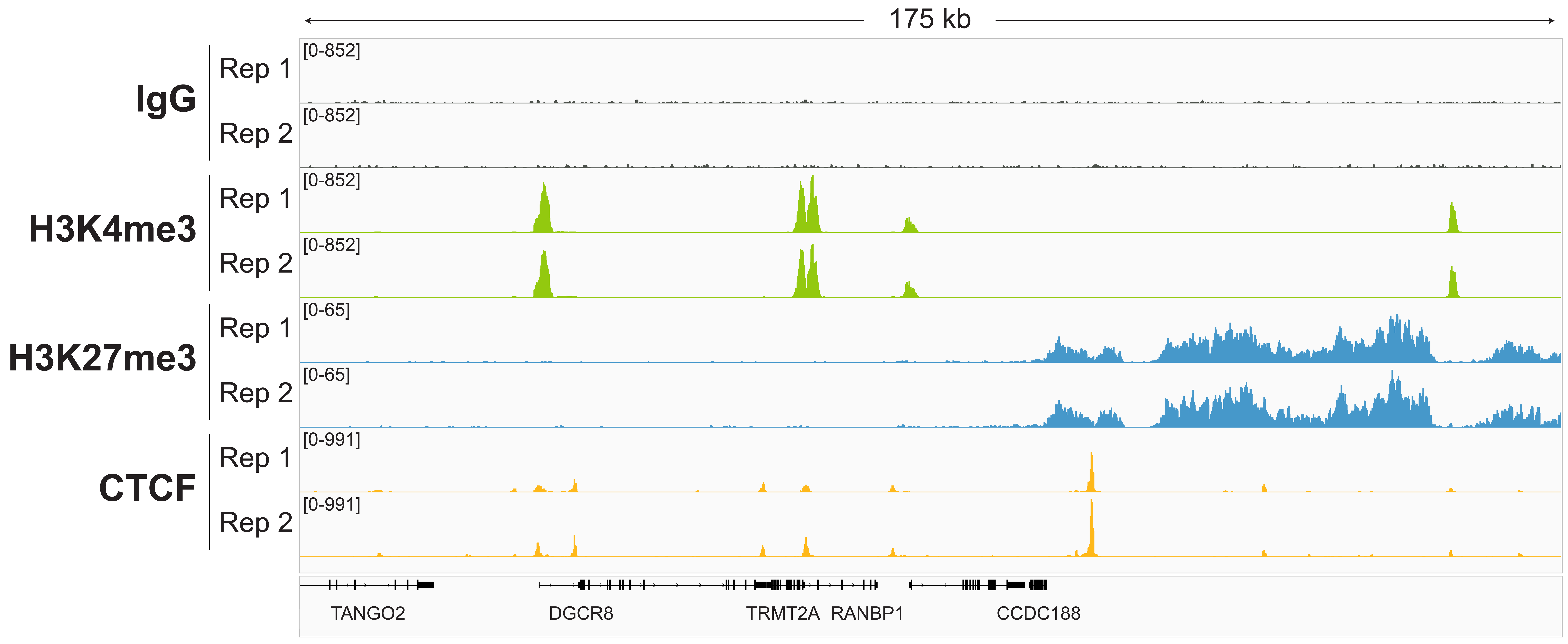
Figure 4: Representative gene browser
tracks
CUT&RUN was performed as described in Figure 5. A representative 175 kb window at the TRMT2A gene is
shown for two replicates (“Rep”) of IgG, H3K4me3, and H3K27me3 kit control antibodies.
Representative tracks are also shown for the transcription factor CTCF. The CUT&RUN kit produced the
expected genomic distribution for each target. Images were generated using the Integrative Genomics
Viewer (IGV, Broad Institute).
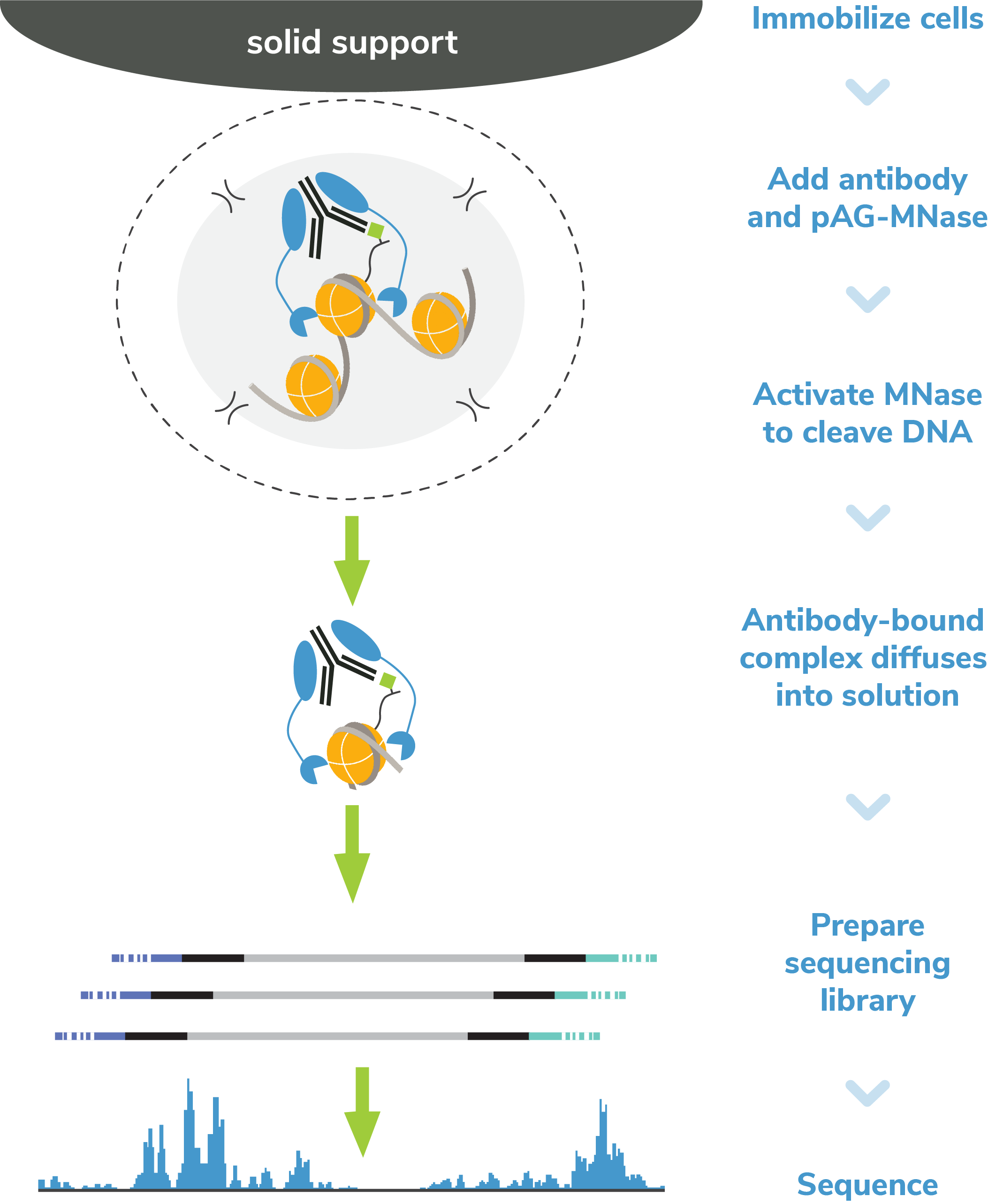
Figure 5: CUT&RUN methods
CUT&RUN was performed using the CUTANA™ ChIC/CUT&RUN Kit starting with 500k K562 cells with 0.5 µg
of IgG (EpiCypher 13-0042), H3K4me3 (EpiCypher 13-0060), H3K27me3 (EpiCypher 13-0055), or 0.125 µg
of CTCF (EpiCypher 13-2014) antibodies in duplicate. Library preparation was performed with 5 ng
of
DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit
(EpiCypher 14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with
paired-end
sequencing (2x50 bp). Sample sequencing depth was 5.5/18.8 million reads (IgG Rep 1/Rep 2),
14.2/17.0 million reads (H3K4me3 Rep 1/Rep 2), 24.7/18.1 million reads (H3K27me3 Rep 1/Rep 2), and
8.6/5.5 million reads (CTCF Rep 1/Rep 2). Data were aligned to the T2T-CHM13v2.0 genome using
Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List
regions.
Kit Contents
| Item | Cat. No. |
|---|---|
| CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows | 15-1016k |
| H3K4me3 Antibody, SNAP-Certified™ for CUT&RUN | 13-0060k |
| H3K27me3 Antibody, SNAP-Certified™ for CUT&RUN | 13-0055k |
| CUTANA™ Rabbit IgG CUT&RUN Negative Control Antibody | 13-0042k |
| SNAP-CUTANA™ K-MetStat Panel | 19-1002k |
| CUTANA™ Concanavalin A Conjugated Paramagnetic Beads | 21-1401k |
| CUTANA™ E. coli Spike-in DNA | 18-1401k |
| CUTANA™ CUT&RUN 8-Strip 0.2 mL Tubes | 10-0009k |
| Bead Activation Buffer | 21-1001k |
| Pre-Wash Buffer | 21-1002k |
| Stop Buffer | 21-1003k |
| 5% Digitonin | 21-1004k |
| 1 M Spermidine | 21-1005k |
| 0.5 M EDTA | 21-1006k |
| 100 mM Calcium Chloride | 21-1007k |
| 0.1X TE Buffer | 21-1025k |
| SPRIselect reagent manufactured by Beckman Coulter, Inc. | 21-1405k |
Recommended Accessory Products
| Item | Cat. No. |
|---|---|
| CUTANA™ CUT&RUN Library Prep Kit | 14-1001/14-1002 |
| SNAP-CUTANA™ K-MetStat Panel | 19-1002 |
| CUT&RUN Antibodies | See the list |
| Magnetic Separation Rack, 0.2 mL Tubes | 10-0008 |
| Magnetic Separation Rack, 1.5 mL Tubes | 10-0012 |
| CUTANA™ Nuclei Extraction Buffer | 21-1026 |
| CUTANA™ Protease Inhibitor Tablets | 21-1027 |
Technical Information
Additional Information
Beckman Coulter, the stylized logo, and SPRIselect are trademarks or registered trademarks of Beckman Coulter, Inc. in the United States and other countries.
Documents & Resources
Kit Version 5
Intellectual Property
US Pat. No. 7790379, 11885814 and related patents and pending applications