CUTANA™ Stop Buffer
{"url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-stop-buffer","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"id":1362,"bulk_discount_rates":[],"can_purchase":true,"meta_description":"CUTANA™ Stop Buffer is used in CUT&RUN to terminate pAG-MNase activity and prevent over-digestion of released DNA fragments.","category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1362/1254/Picture1__91113.1737784711.jpg?c=2","alt":"CUTANA™ Stop Buffer"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1362","shipping":{"calculated":true},"num_reviews":0,"weight":"0.01 LBS","custom_fields":[{"id":"1340","name":"Pack Size","value":"48 Reactions"}],"sku":"21-1003","description":"<!-- <div class=\"product-general-info\">\n <ul style=\"display: none\" class=\"product-general-info__list-left\">\n <li class=\"product-general-info__list-item\"></li>\n <li class=\"product-general-info__list-item\"></li>\n </ul>\n <ul style=\"display: none\" class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\"></li>\n <li class=\"product-general-info__list-item\"></li>\n </ul>\n <ul style=\"padding: 0\" class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <a href=\"#bioz\">\n <div id=\"w-s-3835-21-1026\" style=\"\n width: max-content;\n height: 58px;\n position: relative;\n overflow-y: hidden;\n \"></div>\n <div id=\"bioz-w-pb-21-1026-div\">\n <a id=\"bioz-w-pb-21-1026\" style=\"font-size: 12px; color: transparent\" href=\"https://www.bioz.com/\"\n target=\"_blank\">\n <img src=\"https://cdn.bioz.com/assets/favicon.png\" style=\"\n width: 11px;\n height: 11px;\n vertical-align: baseline;\n padding-bottom: 0px;\n margin-left: 0px;\n margin-bottom: 0px;\n float: none;\n display: none;\n \" />\n </a>\n </div>\n </a>\n </li>\n </ul>\n</div> -->\n<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <p>\n CUTANA<sup>™</sup> Stop Buffer is used in CUT&RUN to terminate pAG-MNase activity and prevent over-digestion\n of released DNA fragments. The Stop Buffer is added following pAG-MNase incubation, and halts enzyme\n activity by chelating free calcium ions.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/kits/21-1003-distribution-analysis.jpg\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"21-1003-distribution-analysis.jpg\"\n src=\"/content/images/products/kits/21-1003-distribution-analysis.jpg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 1: CUT&RUN DNA fragment size distribution\n analysis</strong><br />\n CUT&RUN was performed as described in <strong>Figure 4</strong>. Library DNA was analyzed by Agilent\n TapeStation®.\n This analysis confirmed that mononucleosomes were predominantly enriched in CUT&RUN (~300 bp peaks\n represent 150 bp nucleosomes + sequencing adapters).\n </span>\n </p>\n </div>\n\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/kits/21-1003-genome-wide-heatmap.jpg\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"21-1003-genome-wide-heatmap.jpg\"\n src=\"/content/images/products/kits/21-1003-genome-wide-heatmap.jpg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 2: CUT&RUN genome-wide heatmaps</strong><br />\n CUT&RUN was performed as described in <strong>Figure 4</strong>. Heatmaps show two replicates (“Rep”) of IgG,\n H3K4me3, and H3K27me3 control antibodies in aligned rows ranked by intensity (top to bottom)\n relative to the H3K4me3 Rep 1 reaction and colored such that red indicates high localized enrichment\n and blue denotes background signal.\n </span>\n </p>\n </div>\n\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/kits/21-1003-representative-gene-browser-tracks.jpg\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img loading=\"lazy\"\n alt=\"21-1003-representative-gene-browser-tracks.jpg\"\n src=\"/content/images/products/kits/21-1003-representative-gene-browser-tracks.jpg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 3: Representative gene browser\n tracks</strong><br />\n CUT&RUN was performed as described in <strong>Figure 4</strong>. A representative 175 kb window at\n the TRMT2A gene is\n shown for two replicates (“Rep”) of IgG, H3K4me3, and H3K27me3 control antibodies. Tracks show the\n expected genomic distribution for each target. Images were generated using the Integrative Genomics\n Viewer (IGV, Broad Institute).\n </span>\n </p>\n </div>\n\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/14-1048-23298006-81-figure-5.png\"\n target=\"_blank\" class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"cut and run methods\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/14-1048-23298006-81-figure-5.png\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 4: CUT&RUN methods</strong><br />\n CUT&RUN was performed using the CUTANA™ ChIC/CUT&RUN Kit starting with 500k K562 cells with 0.5 µg\n of IgG (EpiCypher <a\n href=\"/products/nucleosomes/snap-cutana-spike-in-controls/cutana-igg-negative-control-antibody-for-cut-run-and-cut-tag\"\n target=\"_blank\">13-0042</a>), H3K4me3 (EpiCypher <a\n href=\"/products/antibodies/h3k4me3-antibody-snap-certified-for-cut-run-and-cut-tag\"\n target=\"_blank\">13-0060</a>), H3K27me3 (EpiCypher <a\n href=\"/products/antibodies/h3k27me3-antibody-snap-certified-for-cut-run-and-cut-and-tag\"\n target=\"_blank\">13-0055</a>), or\n 0.125 µg\n of CTCF (EpiCypher <a\n href=\"/products/antibodies/cutana-cut-run-compatible-antibodies/ctcf-cutana-cut-and-run-antibody\"\n target=\"_blank\">13-2014</a>) antibodies in duplicate. Library\n preparation was performed with 5 ng of\n DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit\n (EpiCypher <a href=\"/products/epigenetics-kits-and-reagents/cutana-cut-run-library-prep-kit\"\n target=\"_blank\">14-1001/14-1002</a>). Libraries were run on an Illumina\n NextSeq2000 with paired-end\n sequencing (2x50 bp). Sample sequencing depth was 5.5/18.8 million reads (IgG Rep 1/Rep 2),\n 14.2/17.0 million reads (H3K4me3 Rep 1/Rep 2), 24.7/18.1 million reads (H3K27me3 Rep 1/Rep 2), and\n 8.6/5.5 million reads (CTCF Rep 1/Rep 2). Data were aligned to the T2T-CHM13v2.0 genome using\n Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List\n regions.\n </span>\n </p>\n </div>\n\n <!-- <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/nucleosomes/14_1001_distribution_analysis.jpg\"\n target=\"_new\"\n class=\"image-picker__main-image-link\"\n ><img\n loading=\"lazy\"\n alt=\"14_1001_distribution_analysis\"\n src=\"/content/images/products/nucleosomes/14_1001_distribution_analysis.jpg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong\n >Figure 2: CUT&RUN DNA Fragment Size Distribution\n Analysis</strong\n ><br />\n CUT&RUN was performed using the CUTANA ChIC/CUT&RUN Kit\n (EpiCypher\n <a\n href=\"/products/epigenetics-reagents-and-assays/cutana-chic-cut-and-run-kit\"\n >14-1048</a\n >) starting with 500k K562 cells. Five nanograms of CUT&RUN\n output DNA from reactions utilizing IgG (EpiCypher\n <a\n href=\"/products/nucleosomes/snap-cutana-spike-in-controls/cutana-rabbit-igg-cut-run-negative-control-antibody\"\n >13-0042</a\n >), H3K4me3 (EpiCypher\n <a\n href=\"/products/antibodies/snap-chip-certified-antibodies/histone-h3k4me3-antibody-snap-chip-certified-cutana-cut-run-compatible\"\n >13-0041</a\n >), H3K27me3 (ABclonal A16199), and CTCF (EpiCypher\n <a\n href=\"/products/antibodies/cutana-cut-run-compatible-antibodies/ctcf-cutana-cut-and-run-antibody\"\n >13-2014</a\n >) antibodies were prepared for paired-end Illumina<sup\n >®</sup\n >\n sequencing with the CUTANA CUT&RUN Library Prep Kit. Library\n DNA was analyzed by Agilent TapeStation<sup>®</sup>, which\n confirmed that mononucleosomes were predominantly enriched\n in CUT&RUN (~300 bp peaks represent 150 bp nucleosomes +\n sequencing adapters). Peaks at ~380 bp correspond to the\n SNAP-CUTANA™ K-MetStat Panel of spike-in controls (EpiCypher\n <a href=\"/products/nucleosomes/snap-cutana-k-metstat-panel\"\n >19-1002</a\n >).\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/nucleosomes/14_1001_target_compatibility.jpg\"\n target=\"_new\"\n class=\"image-picker__main-image-link\">\n <img\n loading=\"lazy\"\n alt=\"14_1001_target_compatibility\"\n src=\"/content/images/products/nucleosomes/14_1001_target_compatibility.jpg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n >\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 3: Representative gene browser tracks</strong\n ><br />\n\n CUT&RUN sequencing libraries described in Figure 2 were\n sequenced on an Illumina<sup>®</sup> NextSeq 2000 with a P3\n cartridge (paired-end 2x50 cycle). Data were aligned to the\n hg19 genome using Bowtie2. Data were filtered to remove\n duplicates, multi-aligned reads, and blacklist regions. A\n representative 640 kb window at the SEPTIN5 gene is shown\n for three replicates (\"Rep\") of IgG and H3K4me3 antibodies,\n as well as individual tracks for H3K27me3 and the\n transcription factor CTCF, demonstrating the robustness and\n reproducibility of the workflow with a variety of targets.\n Sequencing libraries prepared with the CUTANA CUT&RUN\n Library Prep kit produced the expected genomic distribution\n for each target. Sample sequencing depth was as follows\n (millions of reads): IgG Rep 1 (20.1), IgG Rep 2 (16.6), IgG\n Rep 3 (16.1), H3K4me3 Rep 1 (7.3), H3K4me3 Rep 2 (17.2),\n H3K4me3 Rep 3 (13.8), H3K27me3 (13.4), CTCF (16.2). Images\n were generated using the Integrative Genomics Viewer (IGV,\n Broad Institute).\n </span>\n </p>\n </div> -->\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img loading=\"lazy\" alt=\"21-1003-distribution-analysis.jpg\"\n src=\"/content/images/products/kits/21-1003-distribution-analysis.jpg\" width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\" role=\"button\" />\n <img loading=\"lazy\" alt=\"21-1003-genome-wide-heatmap\"\n src=\"/content/images/products/kits/21-1003-genome-wide-heatmap.jpg\" class=\"image-picker__side-image\"\n role=\"button\" />\n <img loading=\"lazy\" alt=\"representative gene browser tracks\"\n src=\"/content/images/products/kits/21-1003-representative-gene-browser-tracks.jpg\"\n class=\"image-picker__side-image\" role=\"button\" />\n <img loading=\"lazy\" alt=\"cut and run methods\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/14-1048-23298006-81-figure-5.png\"\n class=\"image-picker__side-image\" role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Recommended Accessory Products</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <table class=\"epicypher-table\">\n <tr>\n <th>Item</th>\n <th>Cat. No.</th>\n </tr>\n\n <tr>\n <td>CUTANA<sup>™</sup> ChIC/CUT&RUN Kit </td>\n <td>\n <a href=\"/products/epigenetics-kits-and-reagents/cutana-chic-cut-run-kit\"\n style=\"text-decoration: none;\">\n 14-1048/14-1048-24\n </a>\n </td>\n </tr>\n\n <tr>\n <td>CUTANA<sup>™</sup> CUT&RUN Library Prep Kit </td>\n <td>\n <a href=\"/products/epigenetics-kits-and-reagents/cutana-cut-run-library-prep-kit\"\n style=\"text-decoration: none;\">\n 14-1001/14-1002\n </a>\n </td>\n </tr>\n\n <tr>\n <td>CUTANA<sup>™</sup> pAG-MNase for ChIC/CUT&RUN </td>\n <td>\n <a style=\"text-decoration: none;\"\n href=\"/products/epigenetics-reagents-and-assays/cutana-pag-mnase-for-chic-cut-and-run-workflows\">\n 15-1016/15-1116\n </a>\n </td>\n </tr>\n\n <tr>\n <td id=\"specific-row\">CUTANA<sup>™</sup> Concanavalin A Conjugated Paramagnetic Beads</td>\n <td>\n <a style=\"text-decoration: none;\"\n href=\"/products/epigenetics-kits-and-reagents/cutana-chic-cut-run-assays/cutana-concanavalin-a-conjugated-paramagnetic-beads\">\n 21-1401/21-1411\n </a>\n </td>\n </tr>\n\n <tr>\n <td id=\"specific-row\">CUTANA<sup>™</sup> Nuclei Extraction Buffer</td>\n <td>\n <a style=\"text-decoration: none;\"\n href=\"/products/epigenetics-kits-and-reagents/cutana-nuclei-extraction-buffer\">\n 21-1026\n </a>\n </td>\n </tr>\n\n <tr>\n <td id=\"specific-row\">CUTANA<sup>™</sup> <em>E. coli</em> Spike-in DNA</td>\n <td>\n <a style=\"text-decoration: none;\"\n href=\"/products/nucleosomes/snap-cutana-spike-in-controls/cutana-e-coli-spike-in-dna\">\n 18-1401\n </a>\n </td>\n </tr>\n\n <tr>\n <td id=\"specific-row\">Magnetic Separation Rack, 0.2 mL/1.5 mL Tubes</td>\n <td>\n <a style=\"text-decoration: none;\"\n href=\"/products/epigenetics-reagents-and-assays/cutana-chic-cut-run-assays/magnetic-separation-rack-0-2-ml-tubes\">\n 10-0008</a>/<a style=\"text-decoration: none;\"\n href=\"/products/epigenetics-reagents-and-assays/magnetic-separation-rack-1-5-ml-tubes\">10-0012</a>\n </td>\n </tr>\n\n <tr>\n <td id=\"specific-row\">CUTANA<sup>™</sup> CUT&RUN 8-strip 0.2 mL Tubes</td>\n <td>\n <a style=\"text-decoration: none;\"\n href=\"/products/epigenetics-kits-and-reagents/cutana-chic-cut-run-assays/cutana-cut-run-8-strip-0-2-ml-tubes\">\n 10-0009\n </a>\n </td>\n </tr>\n </table>\n </div>\n </div>\n </div>\n\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Stable for 12 months at 4°C from date of receipt.\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Formulation</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 50 µg/mL RNase A, 50 µg/mL glycogen.\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Instructions for Use</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n CUTANA™ Stop Buffer includes sufficient buffer for 48 reactions using the CUTANA™ CUT&RUN protocol (<a\n href=\"/protocols\">www.epicypher.com/protocols</a>).\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n\n\n <!-- <h3\n style=\"color: #4698cb; font-size: 24px; border-bottom: none\"\n class=\"custom-title-description\">\n Documents & Resources\n </h3> -->\n\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <!-- <h3 style=\"color: #4698cb; margin-bottom: 0\">Current Lot</h3> -->\n <h3 class=\"sub-title1\">Documents & Resources </h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-documents\">\n <a href=\"/content/documents/tds/21-1003.pdf\" target=\"_blank\" class=\"product-documents__link\">\n <svg version=\"1.1\" id=\"Layer_1\" xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\" x=\"0px\" y=\"0px\" viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\" xml:space=\"preserve\" class=\"product-documents__icon\"\n alt=\"16-0002 Datasheet\">\n <g>\n <path class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect x=\"20\" y=\"112\" class=\"product-documents__svg-background\" width=\"146\" height=\"76\" />\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Technical Datasheet</span>\n </a>\n </div>\n </div>\n </div>\n </div>\n\n </div>\n</div>\n\n<!-- <script>\n $(document).ready(function () {\n var widget_micro_obj = new v_widget_obj('s', [1]);\n widget_micro_obj.request_catalog_number_widget_data_internal(\n '21-1026',\n '21-1026'\n );\n });\n</script> -->\n\n<style>\n .form-field-title .required-text {\n display: none !important;\n }\n\n .form-label {\n padding-left: 2rem;\n }\n\n .form-label-text {\n margin: 0;\n margin-left: 0 !important;\n }\n\n /* //////////// */\n\n /* BIOZ */\n td table {\n margin: 0;\n padding: 6px !important;\n }\n\n /* span.bioz-w-parent-hover {\n font-size: 15px;\n }\n\n td .bioz-w-tooltipx {\n padding-top: 0 !important;\n } */\n\n /* .bioz-w-parent-hover:hover {\n text-decoration: underline !important;\n } */\n\n /* /////////// */\n\n .form-field-title .required-text {\n display: none !important;\n }\n\n .form-label {\n padding-left: 2rem;\n }\n\n .form-label-text {\n margin: 0;\n margin-left: 0 !important;\n }\n\n /* .bioz-w-header td {\n padding: 0;\n } */\n\n .epicypher-table table {\n margin-right: auto;\n margin-left: auto;\n width: 100%;\n border-top: 1px solid lightgray;\n }\n\n .epicypher-table th {\n color: #3b3a48;\n }\n\n .epicypher-table td {\n color: #3b3a48;\n font-size: 0.9rem;\n }\n\n .epicypher-table td a {\n text-decoration: underline;\n }\n\n .epicypher-table td,\n th {\n border: 0.5px solid lightgray;\n text-align: left;\n padding: 8px;\n /* white-space: nowrap; */\n }\n\n .epicypher-table tr:nth-child(even) {\n background-color: #f9f9f9;\n }\n\n .epicypher-table td:first-child {\n border-left: 1px solid lightgray;\n }\n\n /* Tablet */\n\n @media only screen and (max-width: 1024px) {\n .epicypher-table th {\n display: none !important;\n }\n\n /* BIOZ */\n /* td.bioz-w-tooltipx::before {\n display: none;\n } */\n\n /* Hide table headers (but not display: none;, for accessibility) */\n .epicypher-table thead tr {\n position: absolute;\n top: -9999px;\n left: -9999px;\n }\n\n .epicypher-table tr {\n border: 1px solid #ccc;\n }\n\n .epicypher-table td {\n /* Behave like a \"row\" */\n /* border: none; */\n border-bottom: 1px solid #eee;\n position: relative;\n padding-left: 30%;\n white-space: inherit;\n }\n\n .epicypher-table td:before {\n /* Now like a table header */\n position: absolute;\n /* Top/left values mimic padding */\n top: 6px;\n left: 6px;\n width: 45%;\n padding-right: 10px;\n white-space: nowrap;\n font-weight: 700;\n }\n\n /*\n Label the data\n */\n .epicypher-table td:nth-of-type(1):before {\n content: 'Item';\n }\n\n td:nth-of-type(2):before {\n content: 'Cat. No.';\n }\n }\n\n @media screen and (min-width: 768px) {\n\n .products-featured .container,\n .products-featured .product-tabs,\n .products-related .container,\n .products-related .product-tabs {\n padding-left: 30px !important;\n padding-right: 50px;\n max-width: 1170px !important;\n }\n\n /* .bioz-w-header,\n .bioz-w-parent-hover td table tbody tr {\n display: inherit;\n } */\n }\n\n @media screen and (max-width: 767px) {\n .custom-title-description {\n font-size: 19px !important;\n }\n\n /* .bioz-w-header,\n .bioz-w-parent-hover td table tbody tr {\n display: flex;\n flex-direction: row;\n } */\n\n .section-title {\n font-size: 19px !important;\n padding-left: 1rem;\n }\n }\n</style>","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$145.00","value":145,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"CUTANA™ Stop Buffer","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"options":[],"related_products":[{"id":1121,"sku":"21-1026","name":"CUTANA™ Nuclei Extraction Buffer","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-nuclei-extraction-buffer","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n \n \n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1121/1183/Screen_Shot_2020-02-12_at_11__54081.1697176206.jpg?c=2","alt":"CUTANA™ Nuclei Extraction Buffer"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1121/1183/Screen_Shot_2020-02-12_at_11__54081.1697176206.jpg?c=2","alt":"CUTANA™ Nuclei Extraction Buffer"}],"date_added":"22nd Sep 2023","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1283,"name":"Pack Size","value":"100 Reactions"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=1121","price":{"without_tax":{"currency":"USD","formatted":"$295.00","value":295},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1121"},{"id":777,"sku":"21-1001","name":"CUTANA™ Bead Activation Buffer","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-bead-activation-buffer","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays"],"summary":"","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/777/1253/Picture1__19497.1737643928.jpg?c=2","alt":"CUTANA™ Bead Activation Buffer"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/777/1253/Picture1__19497.1737643928.jpg?c=2","alt":"CUTANA™ Bead Activation Buffer"}],"date_added":"13th Aug 2020","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1339,"name":"Pack Size","value":"48 Reactions"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=777","price":{"without_tax":{"currency":"USD","formatted":"$145.00","value":145},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=777"},{"id":1363,"sku":"21-1004","name":"CUTANA™ 5% Digitonin","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-digitonin","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays"],"summary":"","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1363/1255/Picture1__22337.1737801428.jpg?c=2","alt":"CUTANA™ 5% Digitonin"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1363/1255/Picture1__22337.1737801428.jpg?c=2","alt":"CUTANA™ 5% Digitonin"}],"date_added":"25th Jan 2025","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1341,"name":"Pack Size","value":"1 mL"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=1363","price":{"without_tax":{"currency":"USD","formatted":"$145.00","value":145},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1363"},{"id":986,"sku":null,"name":"CUTANA™ CUT&Tag Kit","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-cut-and-tag-kit","availability":"","rating":null,"brand":{"name":null},"category":["Antibodies/CUTANA™ CUT&Tag Antibodies","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/986/1213/CTK20__46720.1713641110.png?c=2","alt":"CUTANA™ CUT&Tag Kit"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/986/1213/CTK20__46720.1713641110.png?c=2","alt":"CUTANA™ CUT&Tag Kit"}],"date_added":"8th Mar 2023","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1219,"name":"Pack Size","value":"48 Reactions"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$2,695.00","value":2695},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=986"},{"id":1228,"sku":null,"name":"CUTANA™ Protease Inhibitor Tablets","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-protease-inhibitor-tablets","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n \n Description\n \n \n This EDTA-free cocktail inhibits various serine and cystein","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1228/1251/Product_image_square_1__66866.1732040858.jpg?c=2","alt":"CUTANA™ Protease Inhibitor Tablets"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1228/1251/Product_image_square_1__66866.1732040858.jpg?c=2","alt":"CUTANA™ Protease Inhibitor Tablets"}],"date_added":"5th Nov 2024","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":null,"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$235.00","value":235},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$235.00","value":235},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$505.00","value":505},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1228"}],"shipping_messages":[],"rating":0,"meta_keywords":"cut&run, cut&run kit, stop buffer, cut&run stop buffer","show_quantity_input":1,"title":"CUTANA™ Stop Buffer","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1362/1254/Picture1__91113.1737784711.jpg?c=2","alt":"CUTANA™ Stop Buffer"}]} Pack Size: 48 Reactions
Description
CUTANA™ Stop Buffer is used in CUT&RUN to terminate pAG-MNase activity and prevent over-digestion of released DNA fragments. The Stop Buffer is added following pAG-MNase incubation, and halts enzyme activity by chelating free calcium ions.
Validation Data
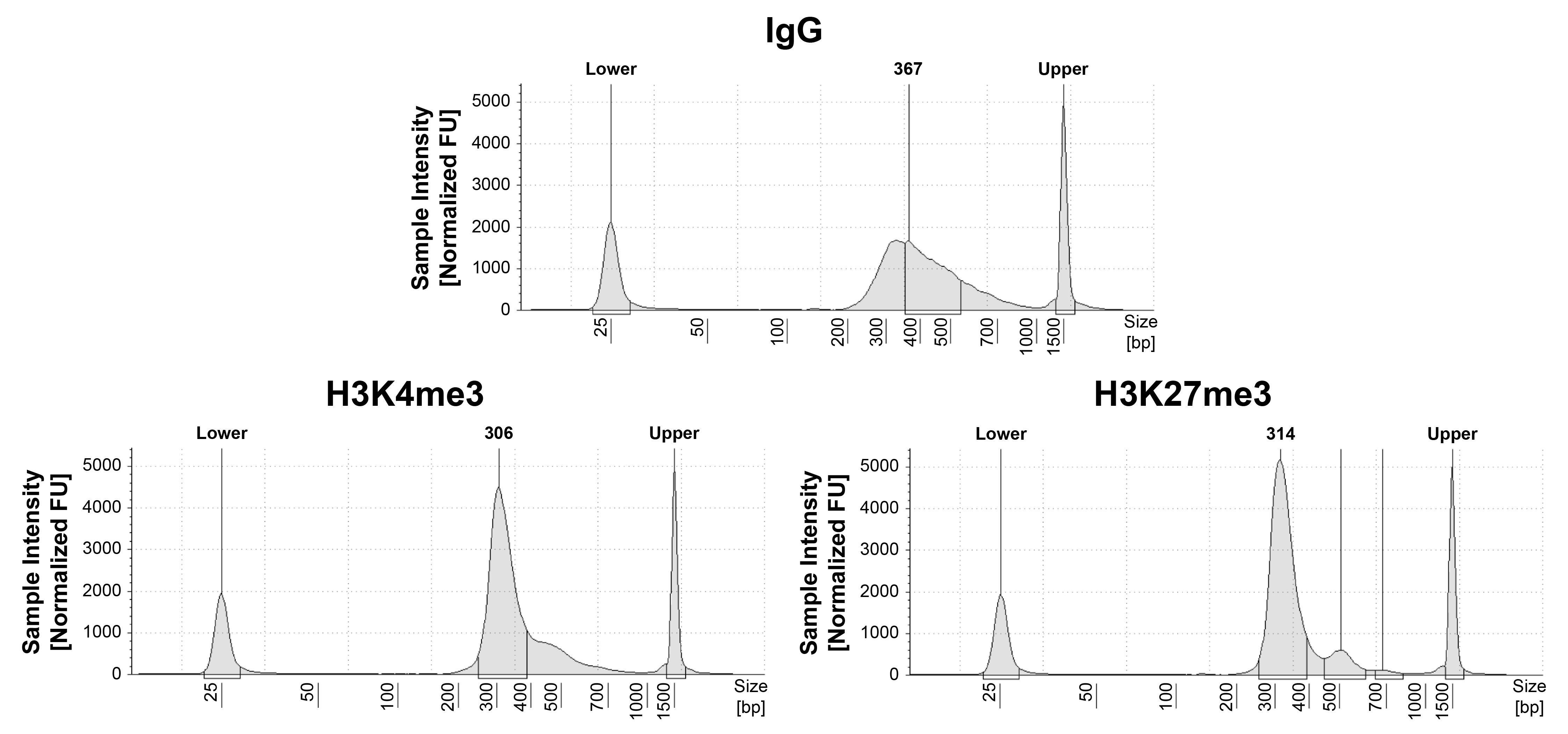
Figure 1: CUT&RUN DNA fragment size distribution
analysis
CUT&RUN was performed as described in Figure 4. Library DNA was analyzed by Agilent
TapeStation®.
This analysis confirmed that mononucleosomes were predominantly enriched in CUT&RUN (~300 bp peaks
represent 150 bp nucleosomes + sequencing adapters).
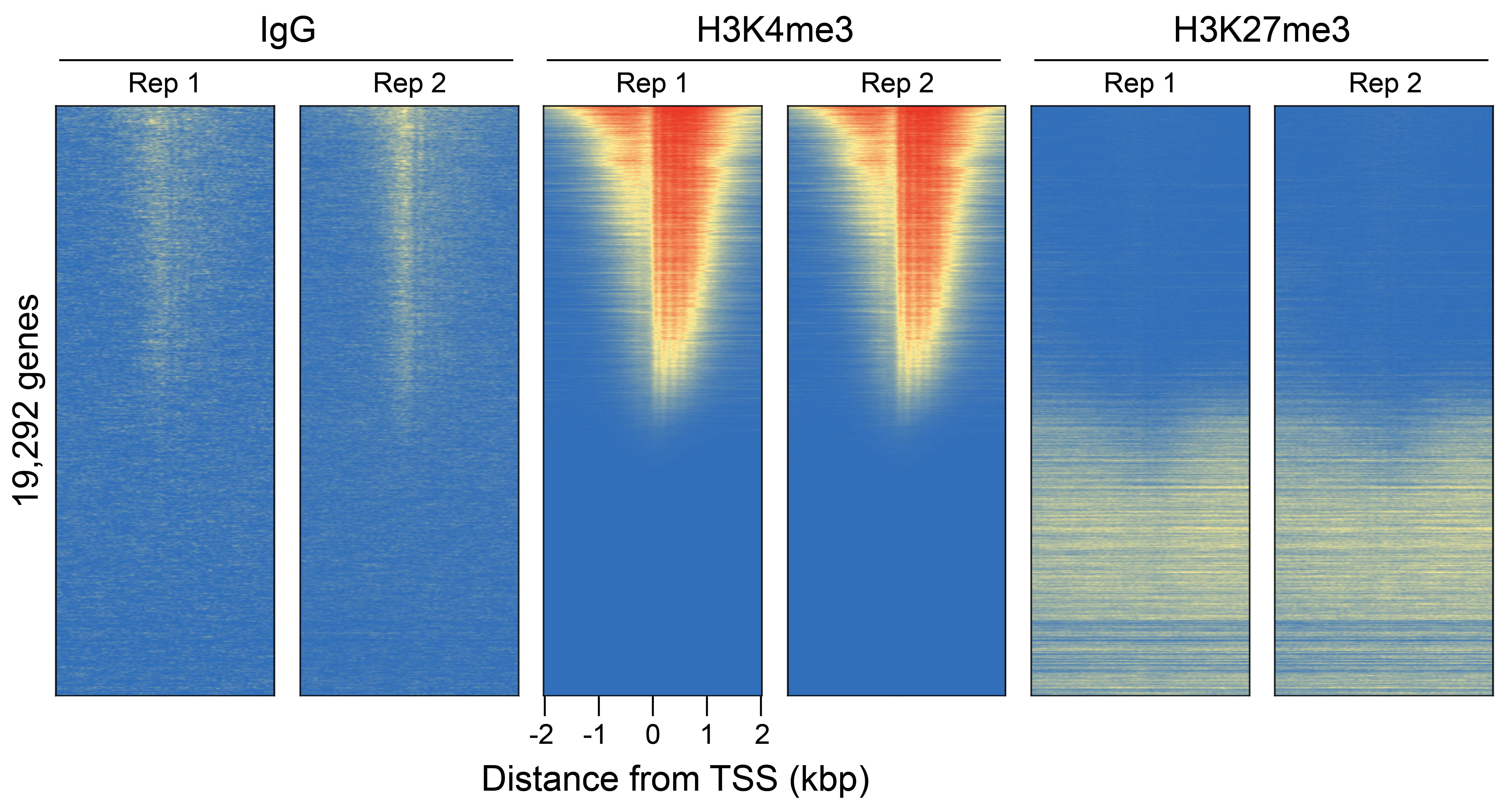
Figure 2: CUT&RUN genome-wide heatmaps
CUT&RUN was performed as described in Figure 4. Heatmaps show two replicates (“Rep”) of IgG,
H3K4me3, and H3K27me3 control antibodies in aligned rows ranked by intensity (top to bottom)
relative to the H3K4me3 Rep 1 reaction and colored such that red indicates high localized enrichment
and blue denotes background signal.
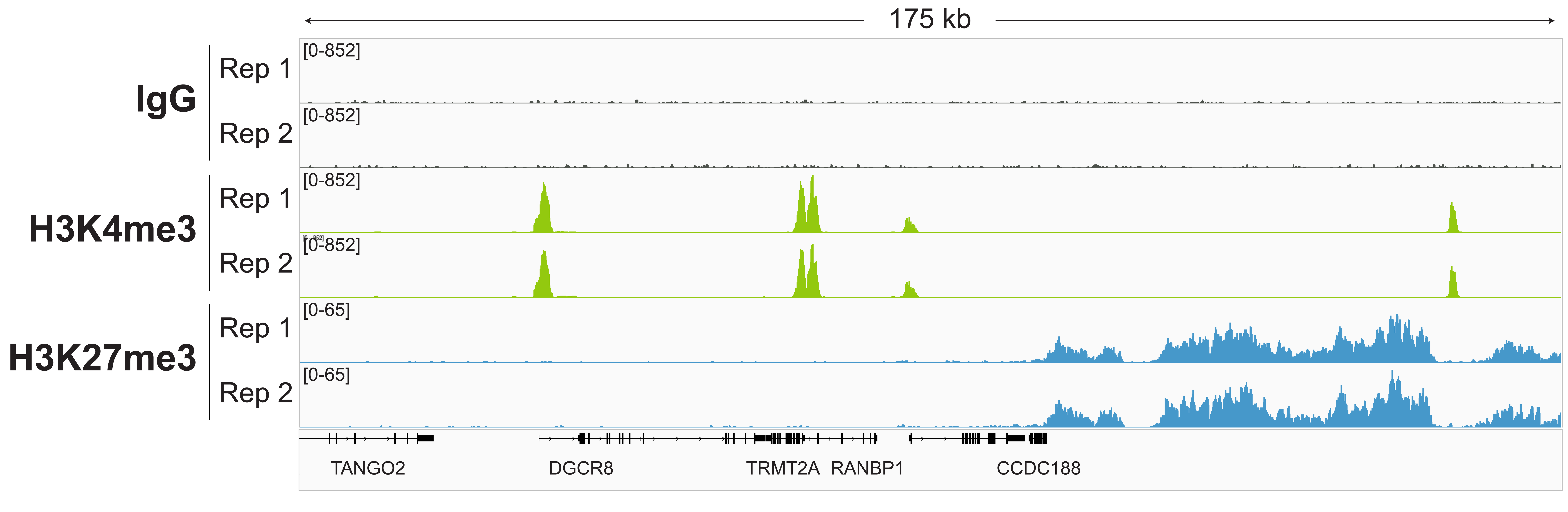
Figure 3: Representative gene browser
tracks
CUT&RUN was performed as described in Figure 4. A representative 175 kb window at
the TRMT2A gene is
shown for two replicates (“Rep”) of IgG, H3K4me3, and H3K27me3 control antibodies. Tracks show the
expected genomic distribution for each target. Images were generated using the Integrative Genomics
Viewer (IGV, Broad Institute).
Figure 4: CUT&RUN methods
CUT&RUN was performed using the CUTANA™ ChIC/CUT&RUN Kit starting with 500k K562 cells with 0.5 µg
of IgG (EpiCypher 13-0042), H3K4me3 (EpiCypher 13-0060), H3K27me3 (EpiCypher 13-0055), or
0.125 µg
of CTCF (EpiCypher 13-2014) antibodies in duplicate. Library
preparation was performed with 5 ng of
DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit
(EpiCypher 14-1001/14-1002). Libraries were run on an Illumina
NextSeq2000 with paired-end
sequencing (2x50 bp). Sample sequencing depth was 5.5/18.8 million reads (IgG Rep 1/Rep 2),
14.2/17.0 million reads (H3K4me3 Rep 1/Rep 2), 24.7/18.1 million reads (H3K27me3 Rep 1/Rep 2), and
8.6/5.5 million reads (CTCF Rep 1/Rep 2). Data were aligned to the T2T-CHM13v2.0 genome using
Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List
regions.
Recommended Accessory Products
| Item | Cat. No. |
|---|---|
| CUTANA™ ChIC/CUT&RUN Kit | 14-1048/14-1048-24 |
| CUTANA™ CUT&RUN Library Prep Kit | 14-1001/14-1002 |
| CUTANA™ pAG-MNase for ChIC/CUT&RUN | 15-1016/15-1116 |
| CUTANA™ Concanavalin A Conjugated Paramagnetic Beads | 21-1401/21-1411 |
| CUTANA™ Nuclei Extraction Buffer | 21-1026 |
| CUTANA™ E. coli Spike-in DNA | 18-1401 |
| Magnetic Separation Rack, 0.2 mL/1.5 mL Tubes | 10-0008/10-0012 |
| CUTANA™ CUT&RUN 8-strip 0.2 mL Tubes | 10-0009 |