CUTANA™ Multiomic CUT&RUN Controls Set
{"url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-multiomic-cut-run-controls-set","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"id":1375,"bulk_discount_rates":[],"can_purchase":true,"meta_description":"CUTANA™ ChIC/CUT&RUN Kit includes all needed reagents to go from cells to purified CUT&RUN DNA. ","category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1375/1265/Multiomic_CR_Icon_Product_Page__02965.1744871009.png?c=2","alt":"CUTANA™ Multiomic CUT&RUN Controls Set"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1375","shipping":{"calculated":true},"num_reviews":0,"weight":"0.01 LBS","custom_fields":[{"id":"1350","name":"Pack Size","value":"1 Set"}],"sku":"14-1802","description":"<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <p>\n The CUTANA™ Multiomic CUT&RUN Controls Set is a specialized set of controls designed to be paired with the\n CUTANA™ ChIC/CUT&RUN Kit (EpiCypher <a href=\"/products/epigenetics-kits-and-reagents/cutana-chic-cut-run-kit\" target=\"_blank\">14-1048</a>) to enable direct, simultaneous\n analysis of DNA methylation and\n chromatin proteins in a single workflow.\n </p>\n \n <p\n style=\"\n background-color: #4698cb;\n color: #fff;\n padding: 1.3rem;\n text-align: center;\n border-radius: 12px;\n \"\n >\n <a\n target=\"_blank\"\n style=\"color: #fff\"\n href=\"/products/epigenetics-kits-and-reagents/cutana-chic-cut-run-kit\"\n >Save 10% when you buy with our 24 Reaction CUT&RUN Kit (14-1048-24)! Learn more >>\n </a>\n </p>\n\n <p>\n Following isolation of chromatin protein-bound DNA in CUT&RUN, DNA\n fragments are prepared for sequencing with a cytosine conversion strategy such as Enzymatic Methyl-seq (NEB<sup>®</sup>\n EM-seq™, preferred) or bisulfite sequencing to provide base-pair resolution of 5-methylcytosine (5mC). This\n set includes the essential controls for CUT&RUN followed by EM-seq (CUT&RUN-EM). H3K36me3 antibody is a\n positive control that enriches DNA associated with active gene bodies, which contain high levels of DNA\n methylation. H3K36me3 antibody complements the antibodies included in the CUT&RUN kit (H3K4me3, which\n enriches unmethylated promoters, and IgG, which serves as a negative control) to validate the technical\n success of the experimental workflow. Pre-fragmented methylated pUC19 and unmethylated Lambda DNAs serve as\n controls in EM-seq to assess conversion efficiency of unmethylated cytosines.\n </p>\n <p>\n This controls set is a tailored solution to unlock the capacity of CUT&RUN to generate multiomic insights\n and investigate crosstalk between DNA methylation and chromatin proteins.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/kits/14-1802-dna-fragment-size-distribution-analysis.jpeg\"\n target=\"_blank\" class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"14-1802-dna-fragment-size-distribution-analysis\"\n src=\"/content/images/products/kits/14-1802-dna-fragment-size-distribution-analysis.jpeg\"\n loading=\"lazy\" class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 1: Representative CUT&RUN-EM DNA fragment size\n distribution analysis</strong><br />\n CUT&RUN-EM was performed as described in Figure 6. Library DNA was analyzed by Agilent TapeStation<sup>®</sup>.\n This analysis confirmed that mononucleosomes were predominantly enriched in CUT&RUN-EM (~300 bp\n peaks represent 150 bp nucleosomes+ sequencing adapters). High molecular weight nucleosome laddering is expected \n with EM-seq and does not affect sequencing quality.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/kits/14-1802-control-dna-fragment-size.jpeg\"\n target=\"_blank\" class=\"image-picker__main-image-link\"><img loading=\"lazy\"\n alt=\"14-1802-control-dna-fragment-size\"\n src=\"/content/images/products/kits/14-1802-control-dna-fragment-size.jpeg\"\n class=\"image-picker__main-image\" loading=\"lazy\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 2: Control DNA fragment size</strong><br />\n TapeStation confirms fragmentation of pUC19 (Lane 1, 0.2 ng) and Lambda (Lane 2, 4 ng) control DNAs\n to target size of 200-400 bp.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/kits/14-1802-conversion-efficiency.jpeg\"\n target=\"_blank\" class=\"image-picker__main-image-link\">\n <img loading=\"lazy\" alt=\"14-1802-conversion-efficiency\"\n src=\"/content/images/products/kits/14-1802-conversion-efficiency.jpeg\"\n loading=\"lazy\" class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span>\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 3: Representative EM-seq conversion efficiency</strong><br />\n CUT&RUN-EM was performed as described in Figure 6. Across multiple CUT&RUN targets, methylated pUC19\n control DNA shows >95% methylated CpGs, as expected. Unmethylated Lambda control DNA shows <1% DNA\n methylation, indicating>99% EM-seq conversion efficiency.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/kits/14-1802-methylation-by-target.jpeg\"\n target=\"_blank\" class=\"image-picker__main-image-link\">\n <img loading=\"lazy\" alt=\"14-1802-methylation-by-target\"\n src=\"/content/images/products/kits/14-1802-methylation-by-target.jpeg\"\n loading=\"lazy\" class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span>\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 4: Representative percent methylation by target</strong><br />\n CUT&RUN-EM was performed as described in Figure 6. Global levels of DNA methylation associated with\n each target (IgG, H3K4me3, and H3K36me3) are shown. The H3K36me3 positive control is highly\n associated with DNA methylation at active gene bodies, while H3K4me3 has very low levels of DNA\n methylation at active promoters.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/kits/14-1802-representative-gene-browser-tracks.jpeg\"\n target=\"_blank\" class=\"image-picker__main-image-link\">\n <img loading=\"lazy\" alt=\"14-1802-representative-gene-browser-tracks\"\n src=\"/content/images/products/kits/14-1802-representative-gene-browser-tracks.jpeg\"\n loading=\"lazy\" class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span>\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 5: Representative gene browser tracks</strong><br />\n CUT&RUN-EM was performed as described in Figure 6. A 103 kb window is shown for IgG, H3K4me3, and\n H3K36me3. CUT&RUN-EM produced the expected genomic distribution for each chromatin protein, whereby\n H3K36me3 peaks are highly enriched for methylated DNA (red) in active gene bodies, and H3K4me3 peaks\n are typically unmethylated DNA (blue) at active promoters. A whole genome EM-seq (WGEM) track\n showing genome-wide DNA methylation is shown for reference. Images were generated using the\n Integrative Genomics Viewer (IGV, Broad Institute).\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/cut-run-em-methods.png?t=1745094750\"\n target=\"_blank\" class=\"image-picker__main-image-link\">\n <img loading=\"lazy\" alt=\"cut-run-em-methods\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/cut-run-em-methods.png?t=1745094750\"\n loading=\"lazy\" class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span>\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 6: CUT&RUN-EM methods</strong><br />\n CUT&RUN-EM was performed using the CUTANA™ Multiomic CUT&RUN Controls Set and the CUTANA™\n ChIC/CUT&RUN Kit (EpiCypher <a href=\"/products/epigenetics-kits-and-reagents/cutana-chic-cut-run-kit\" target=\"_blank\">14-1048</a>) starting with 500k K562 cells and 0.5 µg of IgG (EpiCypher\n <a href=\"/products/nucleosomes/snap-cutana-spike-in-controls/cutana-igg-negative-control-antibody-for-cut-run-and-cut-tag\" target=\"_blank\">13-0042)</a>, H3K4me3 (EpiCypher <a href=\"/products/antibodies/h3k4me3-antibody-snap-certified-for-cut-run-and-cut-tag\" target=\"_blank\">13-0060</a>), or H3K36me3 (EpiCypher <a href=\"/products/antibodies/cut-and-run-antibodies/cut-and-run-antibodies-histone-ptms/h3k36me3-antibody-snap-certified-for-cut-and-run\" target=\"_blank\">13-0058</a>) antibodies. Library\n preparation was performed with 1 ng of DNA using the NEBNext<sup>®</sup> Enzymatic Methyl-seq v2 Kit (NEB\n E8015). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2x50 bp). Sample\n sequencing depth was 31.5 million reads (IgG), 21.3 million reads (H3K4me3), and 20.6 million reads\n (H3K36me3). Data were aligned to the hg38 genome using Bowtie2. Data were filtered to remove\n duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions. Validation data are\n representative where noted. Refer to the product page for <a href=\"/products/antibodies/cut-and-run-antibodies/cut-and-run-antibodies-histone-ptms/h3k36me3-antibody-snap-certified-for-cut-and-run\" target=\"_blank\">13-0058</a> for the lot-specific validation\n approach for H3K36me3 antibody.\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img loading=\"lazy\" alt=\"14-1802-dna-fragment-size-distribution-analysis\"\n src=\"/content/images/products/kits/14-1802-dna-fragment-size-distribution-analysis.jpeg\"\n width=\"200\" loading=\"lazy\" class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\" />\n <img loading=\"lazy\" alt=\"14-1802-control-dna-fragment-size\"\n src=\"/content/images/products/kits/14-1802-control-dna-fragment-size.jpeg\"\n class=\"image-picker__side-image\" loading=\"lazy\" role=\"button\" />\n <img loading=\"lazy\" alt=\"14-1802-conversion-efficiency\"\n src=\"/content/images/products/kits/14-1802-conversion-efficiency.jpeg\"\n loading=\"lazy\" class=\"image-picker__side-image\" role=\"button\" />\n <img loading=\"lazy\" alt=\"14-1802-methylation-by-target\"\n src=\"/content/images/products/kits/14-1802-methylation-by-target.jpeg\"\n loading=\"lazy\" class=\"image-picker__side-image\" role=\"button\" />\n <img loading=\"lazy\" alt=\"14-1048-RGBT\"\n src=\"/content/images/products/kits/14-1802-representative-gene-browser-tracks.jpeg\"\n loading=\"lazy\" class=\"image-picker__side-image\" role=\"button\" />\n <img loading=\"lazy\" alt=\"cut-run-em-methods\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/cut-run-em-methods.png?t=1745094750\"\n loading=\"lazy\" class=\"image-picker__side-image\" role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Set Contents</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <table class=\"epicypher-table\">\n <tr>\n <th>Item</th>\n <th>Cat. No.</th>\n <th>QTY</th>\n </tr>\n\n <tr>\n <td>CUTANA™ H3K36me3 Positive Control Antibody</td>\n <td>\n <a\n href=\"/products/antibodies/cut-and-run-antibodies/cut-and-run-antibodies-histone-ptms/h3k36me3-antibody-snap-certified-for-cut-and-run\" target=\"_blank\">13-0058</a>-03\n </td>\n <td>8 rxn</td>\n </tr>\n\n <tr>\n <td>CUTANA™ CpG Methylated pUC19 Fragmented Control DNA</td>\n <td>\n 18-8001-05\n </td>\n <td>20 µL</td>\n </tr>\n\n <tr>\n <td>\n CUTANA™ CpG Unmethylated Lambda Fragmented Control DNA\n </td>\n <td>\n 18-8002-05\n </td>\n <td>20 µL</td>\n </tr>\n\n <tr>\n <td>CUTANA™ 0.1X TE Buffer</td>\n <td>\n 21-1025-05\n </td>\n <td>2 x 2 mL</td>\n </tr>\n </table>\n </div>\n </div>\n </div>\n\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Recommended Accessory Products</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <table class=\"epicypher-table\">\n <tr>\n <th>Item</th>\n <th>Cat. No.</th>\n </tr>\n\n <tr>\n <td>CUTANA™ ChIC/CUT&RUN Kit</td>\n <td>\n <a\n href=\"/products/epigenetics-kits-and-reagents/cutana-chic-cut-run-kit\" target=\"_blank\">14-1048/14-1048-24</a>\n </td>\n </tr>\n\n <tr>\n <td>NEBNext<sup>®</sup> Enzymatic Methyl-seq v2 Kit </td>\n <td>\n NEB E8015\n </td>\n </tr>\n </table>\n </div>\n </div>\n </div>\n\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n OPEN SET IMMEDIATELY and store components at room\n temperature, 4°C, and -20°C as indicated. Stable for 6 months upon date of receipt.\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Instructions for Use</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n See User Manual corresponding to Controls Set Version 1.\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <!-- <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Additional Information</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <p>\n Beckman Coulter, the stylized logo, and SPRIselect are\n trademarks or registered trademarks of Beckman Coulter, Inc.\n in the United States and other countries.\n </p>\n </div>\n </div>\n </div> -->\n\n <h3 style=\"color: #4698cb; font-size: 24px; border-bottom: none\" class=\"custom-title-description\">\n Documents & Resources\n </h3>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <!-- <h3 style=\"color: #4698cb; margin-bottom: 0\">Current Lot</h3> -->\n <h3 style=\"margin-top: 1rem; padding-top: 0\" class=\"sub-title1\">\n Set Controls Version 1\n </h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-documents\">\n <a href=\"/content/documents/tds/14-1802.pdf\" target=\"_blank\" class=\"product-documents__link\">\n <svg version=\"1.1\" id=\"Layer_1\" xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\" x=\"0px\" y=\"0px\" viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\" xml:space=\"preserve\" class=\"product-documents__icon\"\n alt=\"16-0002 Datasheet\">\n <g>\n <path class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect x=\"20\" y=\"112\" class=\"product-documents__svg-background\" width=\"146\" height=\"76\" />\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Technical Datasheet\n </span>\n </a>\n </div>\n <div class=\"product-documents\">\n <a href=\"/content/documents/manuals/multiomic-cut-and-run-manual.pdf\" target=\"_blank\"\n class=\"product-documents__link\">\n <svg version=\"1.1\" id=\"Layer_1\" xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\" x=\"0px\" y=\"0px\" viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\" xml:space=\"preserve\" class=\"product-documents__icon\"\n alt=\"16-0002 Datasheet\">\n <g>\n <path class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect x=\"20\" y=\"112\" class=\"product-documents__svg-background\" width=\"146\" height=\"76\" />\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Multiomic CUT&RUN Controls Set Manual</span>\n </a>\n </div>\n <div class=\"product-documents\">\n <a href=\"/content/documents/manuals/multiomic-cut-and-run-quick-card.pdf\" target=\"_blank\"\n class=\"product-documents__link\">\n <svg version=\"1.1\" id=\"Layer_1\" xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\" x=\"0px\" y=\"0px\" viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\" xml:space=\"preserve\" class=\"product-documents__icon\"\n alt=\"16-0002 Datasheet\">\n <g>\n <path class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect x=\"20\" y=\"112\" class=\"product-documents__svg-background\" width=\"146\" height=\"76\" />\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Multiomic CUT&RUN Controls Set Quick Card</span>\n </a>\n </div>\n </div>\n </div>\n </div>\n </div>\n</div>\n\n<script>\n $(document).ready(function () {\n var widget_micro_obj = new v_widget_obj(\"s\", [1]);\n widget_micro_obj.request_catalog_number_widget_data_internal(\n \"14-1048\",\n \"14-1048\"\n );\n });\n</script>\n\n<style>\n .form-field-title .required-text {\n display: none !important;\n }\n\n .form-label {\n padding-left: 2rem;\n }\n\n .form-label-text {\n margin: 0;\n margin-left: 0 !important;\n }\n\n /* //////////// */\n\n /* BIOZ */\n td table {\n margin: 0;\n padding: 6px !important;\n }\n\n span.bioz-w-parent-hover {\n font-size: 15px;\n }\n\n td .bioz-w-tooltipx {\n padding-top: 0 !important;\n }\n\n /* .bioz-w-parent-hover:hover {\n text-decoration: underline white 2px !important;\n } */\n\n /* /////////// */\n\n .form-field-title .required-text {\n display: none !important;\n }\n\n .form-label {\n padding-left: 2rem;\n }\n\n .form-label-text {\n margin: 0;\n margin-left: 0 !important;\n }\n\n .epicypher-table table {\n margin-right: auto;\n margin-left: auto;\n width: 100%;\n border-top: 1px solid lightgray;\n }\n\n .epicypher-table th {\n color: #3b3a48;\n }\n\n .epicypher-table td {\n color: #3b3a48;\n font-size: 0.9rem;\n }\n\n .epicypher-table td a {\n text-decoration: underline;\n }\n\n .epicypher-table td,\n th {\n border: 0.5px solid lightgray;\n text-align: left;\n padding: 8px;\n /* white-space: nowrap; */\n }\n\n .epicypher-table tr:nth-child(even) {\n background-color: #f9f9f9;\n }\n\n .epicypher-table td:first-child {\n border-left: 1px solid lightgray;\n }\n\n /* Tablet */\n\n @media only screen and (max-width: 1024px) {\n\n /* Force table to not be like tables anymore */\n .epicypher-table th {\n display: none !important;\n }\n\n /* BIOZ */\n td.bioz-w-tooltipx::before {\n display: none;\n }\n\n /* Hide table headers (but not display: none;, for accessibility) */\n .epicypher-table thead tr {\n position: absolute;\n top: -9999px;\n left: -9999px;\n }\n\n .epicypher-table tr {\n border: 1px solid #ccc;\n }\n\n .epicypher-table td {\n /* Behave like a \"row\" */\n /* border: none; */\n border-bottom: 1px solid #eee;\n position: relative;\n padding-left: 30%;\n white-space: inherit;\n }\n\n .epicypher-table td:before {\n /* Now like a table header */\n position: absolute;\n /* Top/left values mimic padding */\n top: 6px;\n left: 6px;\n width: 45%;\n padding-right: 10px;\n white-space: nowrap;\n font-weight: 700;\n }\n\n /*\n Label the data\n */\n .epicypher-table td:nth-of-type(1):before {\n content: \"Item\";\n }\n\n td:nth-of-type(2):before {\n content: \"Cat. No.\";\n }\n }\n\n @media screen and (min-width: 768px) {\n\n .products-featured .container,\n .products-featured .product-tabs,\n .products-related .container,\n .products-related .product-tabs {\n padding-left: 30px !important;\n padding-right: 50px;\n max-width: 1170px !important;\n }\n }\n\n @media screen and (max-width: 767px) {\n .custom-title-description {\n font-size: 19px !important;\n }\n\n .section-title {\n font-size: 19px !important;\n padding-left: 1rem;\n }\n }\n</style>","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$225.00","value":225,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"CUTANA™ Multiomic CUT&RUN Controls Set","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"options":[],"related_products":[{"id":758,"sku":"13-0042","name":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag","url":"https://www.epicypher.com/products/nucleosomes/snap-cutana-spike-in-controls/cutana-igg-negative-control-antibody-for-cut-run-and-cut-tag","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes/SNAP-CUTANA™ Spike-in Controls","Antibodies/CUTANA™ CUT&RUN Antibodies","Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n Type: Polyclonal\n \n \n Host: Rabbit\n \n \n Applications: CUT&R","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/758/739/1579731877.1280.1280__61771.1592491196.png?c=2","alt":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/758/739/1579731877.1280.1280__61771.1592491196.png?c=2","alt":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag"}],"date_added":"17th Jun 2020","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1148,"name":"Pack Size","value":"100 µg"},{"id":1149,"name":"Internal Comment","value":"bulk stocks from Thermo in cold room"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=758","price":{"without_tax":{"currency":"USD","formatted":"$105.00","value":105},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=758"},{"id":917,"sku":"13-2018","name":"EGFR CUTANA™ CUT&RUN Antibody","url":"https://www.epicypher.com/products/antibodies/cutana-cut-run-antibodies/egfr-cutana-cut-run-antibody","availability":"","rating":null,"brand":{"name":null},"category":["Antibodies/CUTANA™ CUT&RUN Antibodies","Antibodies/CUTANA™ CUT&RUN Antibodies/CUT&RUN Antibodies - Chromatin-Associated Proteins","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n Type: Polyclonal\n \n \n Host: Rabbit\n \n \n Applications: CUT&R","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/917/1018/chd3-cutana-cutandrun-antibody__66975.1645734525__91145.1648666822__03424.1651862137__58623.1652127357__88640.1652155700.jpg?c=2","alt":"EGFR CUTANA™ CUT&RUN Antibody"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/917/1018/chd3-cutana-cutandrun-antibody__66975.1645734525__91145.1648666822__03424.1651862137__58623.1652127357__88640.1652155700.jpg?c=2","alt":"EGFR CUTANA™ CUT&RUN Antibody"}],"date_added":"10th May 2022","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1085,"name":"Pack Size","value":"100 µL"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=917","price":{"without_tax":{"currency":"USD","formatted":"$525.00","value":525},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=917"},{"id":874,"sku":"13-2014","name":"CTCF CUTANA™ CUT&RUN Antibody","url":"https://www.epicypher.com/products/antibodies/cutana-cut-run-compatible-antibodies/ctcf-cutana-cut-and-run-antibody","availability":"","rating":null,"brand":{"name":null},"category":["Antibodies/CUTANA™ CUT&RUN Antibodies","Antibodies/CUTANA™ CUT&RUN Antibodies/CUT&RUN Antibodies - Chromatin-Associated Proteins","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n Type: Monoclonal\n \n \n Host: Rabbit\n \n \n Applications: CUT&R","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/874/968/ctcf-cutana-cutandrun-antibody__00748.1645734560.jpg?c=2","alt":"CTCF CUTANA CUTandRUN Antibody"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/874/968/ctcf-cutana-cutandrun-antibody__00748.1645734560.jpg?c=2","alt":"CTCF CUTANA CUTandRUN Antibody"}],"date_added":"20th Jul 2021","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":908,"name":"Pack Size","value":"100 µL"}],"num_reviews":null,"weight":{"formatted":"0.00 LBS","value":0},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=874","price":{"without_tax":{"currency":"USD","formatted":"$525.00","value":525},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=874"},{"id":798,"sku":"13-2003","name":"BRD4 CUTANA™ CUT&RUN Antibody","url":"https://www.epicypher.com/products/antibodies/brd4-cutana-cut-run-antibody","availability":"","rating":null,"brand":{"name":null},"category":["Antibodies","Antibodies/CUTANA™ CUT&RUN Antibodies","Antibodies/CUTANA™ CUT&RUN Antibodies/CUT&RUN Antibodies - Chromatin-Associated Proteins","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n Type: Polyclonal\n \n \n Host: Rabbit\n \n \n Applications: CUT&RUN, IH","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/798/933/brd4-cutana-cutandrun-antibody__13949.1645734524.jpg?c=2","alt":"BRD4 CUTANA CUTandRUN Antibody"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/798/933/brd4-cutana-cutandrun-antibody__13949.1645734524.jpg?c=2","alt":"BRD4 CUTANA CUTandRUN Antibody"}],"date_added":"25th Nov 2020","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":655,"name":"Pack size","value":"50 µL"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=798","price":{"without_tax":{"currency":"USD","formatted":"$525.00","value":525},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=798"},{"id":1232,"sku":null,"name":"CUTANA™ ChIC/CUT&RUN Kit","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-chic-cut-run-kit","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n \n \n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1232/1252/Epicypher_CUTANA_CUTRUN_Box__44424.1734029530.png?c=2","alt":"CUTANA™ ChIC/CUT&RUN Kit"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1232/1252/Epicypher_CUTANA_CUTRUN_Box__44424.1734029530.png?c=2","alt":"CUTANA™ ChIC/CUT&RUN Kit"}],"date_added":"12th Dec 2024","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1337,"name":"Pack size","value":"48 Reactions"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$945.00","value":945},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1232"}],"shipping_messages":[],"rating":0,"meta_keywords":"CUTANA, CUT&RUN, CUT&RUN kit, CUT and RUN, CUT and RUN kit, CUTANA CUT&RUN Kit, pAG-MNase kit, genomic mapping, ChIP-Seq","show_quantity_input":1,"title":"CUTANA™ Multiomic CUT&RUN Controls Set","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1375/1265/Multiomic_CR_Icon_Product_Page__02965.1744871009.png?c=2","alt":"CUTANA™ Multiomic CUT&RUN Controls Set"}]} Pack Size: 1 Set
Description
The CUTANA™ Multiomic CUT&RUN Controls Set is a specialized set of controls designed to be paired with the CUTANA™ ChIC/CUT&RUN Kit (EpiCypher 14-1048) to enable direct, simultaneous analysis of DNA methylation and chromatin proteins in a single workflow.
Save 10% when you buy with our 24 Reaction CUT&RUN Kit (14-1048-24)! Learn more >>
Following isolation of chromatin protein-bound DNA in CUT&RUN, DNA fragments are prepared for sequencing with a cytosine conversion strategy such as Enzymatic Methyl-seq (NEB® EM-seq™, preferred) or bisulfite sequencing to provide base-pair resolution of 5-methylcytosine (5mC). This set includes the essential controls for CUT&RUN followed by EM-seq (CUT&RUN-EM). H3K36me3 antibody is a positive control that enriches DNA associated with active gene bodies, which contain high levels of DNA methylation. H3K36me3 antibody complements the antibodies included in the CUT&RUN kit (H3K4me3, which enriches unmethylated promoters, and IgG, which serves as a negative control) to validate the technical success of the experimental workflow. Pre-fragmented methylated pUC19 and unmethylated Lambda DNAs serve as controls in EM-seq to assess conversion efficiency of unmethylated cytosines.
This controls set is a tailored solution to unlock the capacity of CUT&RUN to generate multiomic insights and investigate crosstalk between DNA methylation and chromatin proteins.
Validation Data
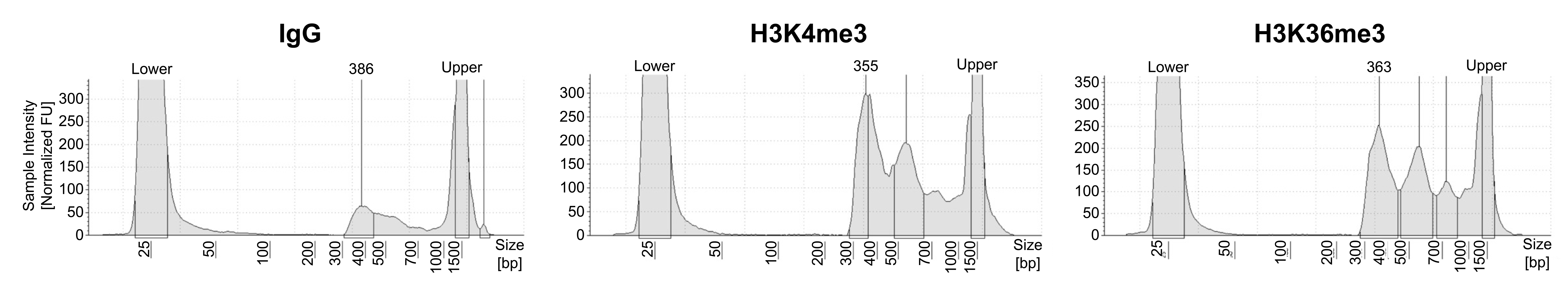
Figure 1: Representative CUT&RUN-EM DNA fragment size
distribution analysis
CUT&RUN-EM was performed as described in Figure 6. Library DNA was analyzed by Agilent TapeStation®.
This analysis confirmed that mononucleosomes were predominantly enriched in CUT&RUN-EM (~300 bp
peaks represent 150 bp nucleosomes+ sequencing adapters). High molecular weight nucleosome laddering is expected
with EM-seq and does not affect sequencing quality.
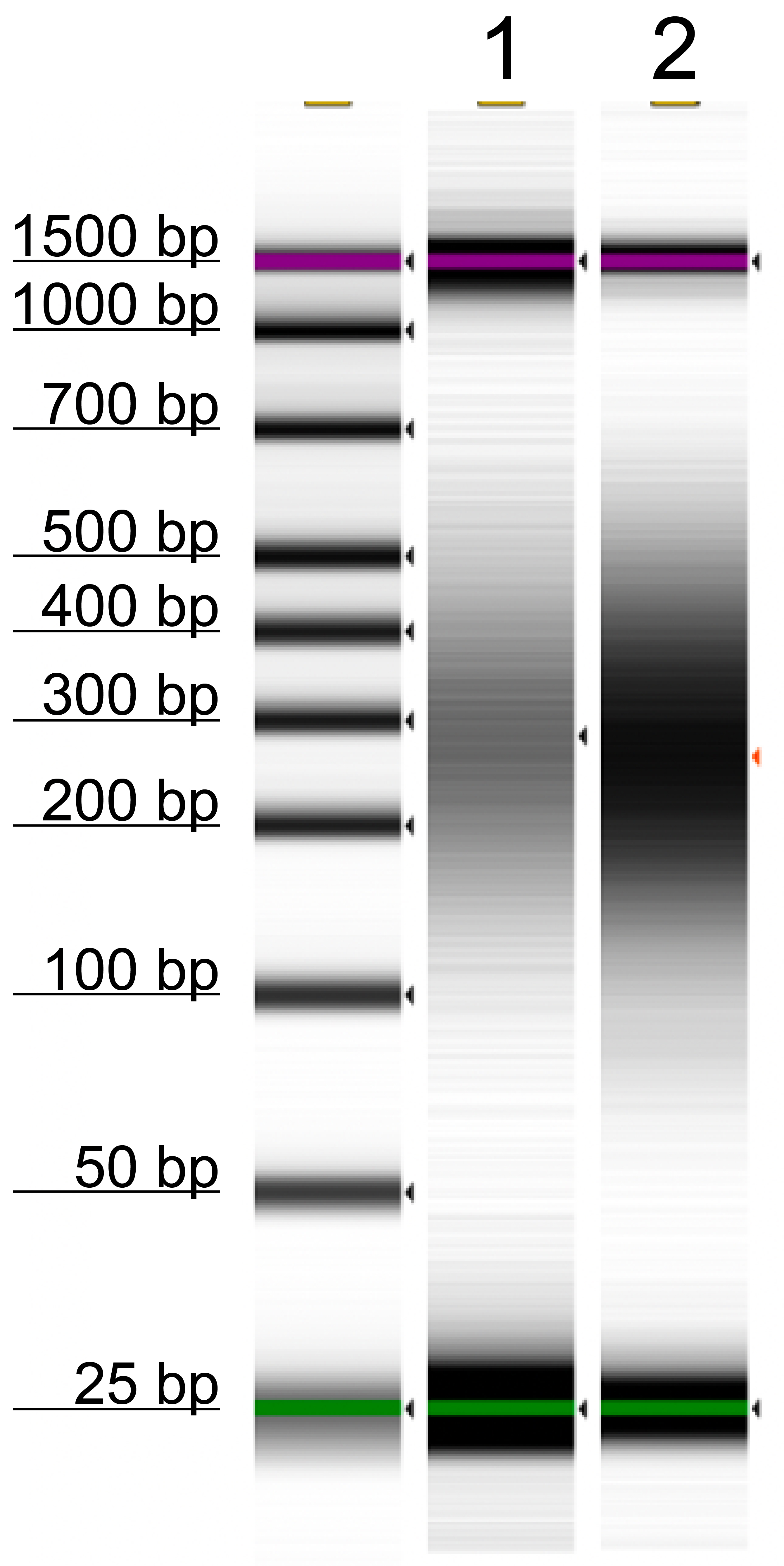
Figure 2: Control DNA fragment size
TapeStation confirms fragmentation of pUC19 (Lane 1, 0.2 ng) and Lambda (Lane 2, 4 ng) control DNAs
to target size of 200-400 bp.
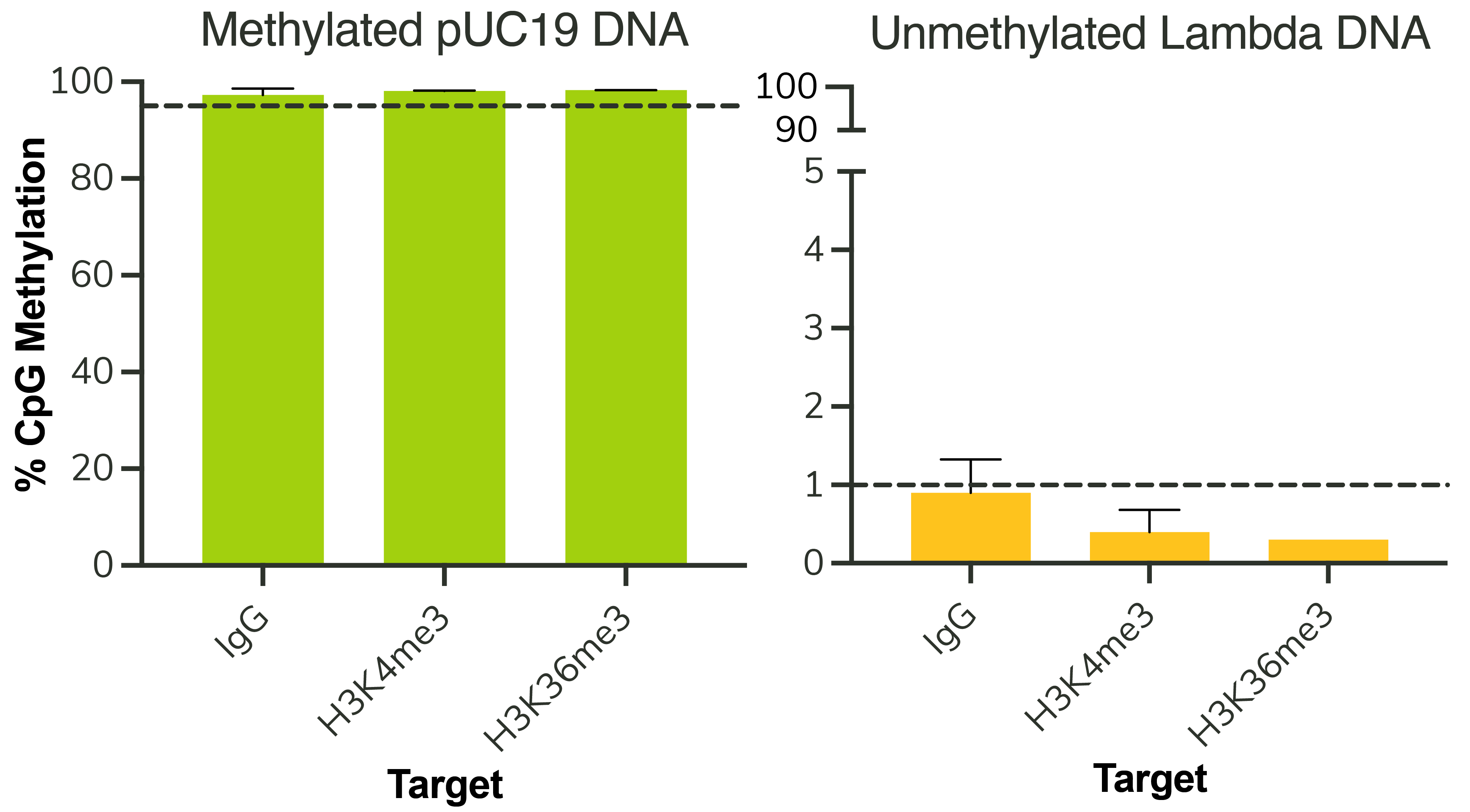
Figure 3: Representative EM-seq conversion efficiency
CUT&RUN-EM was performed as described in Figure 6. Across multiple CUT&RUN targets, methylated pUC19
control DNA shows >95% methylated CpGs, as expected. Unmethylated Lambda control DNA shows <1% DNA
methylation, indicating>99% EM-seq conversion efficiency.
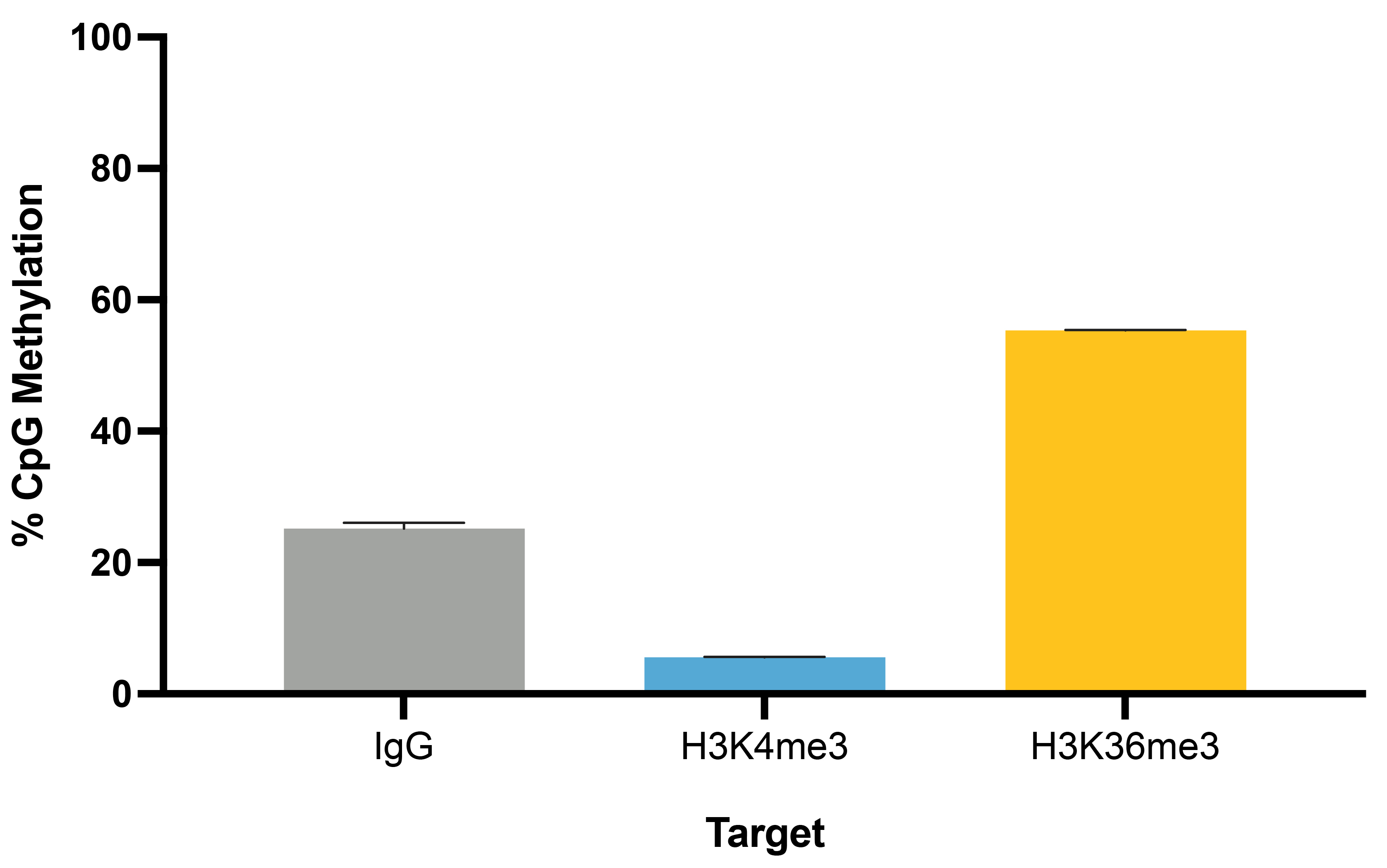
Figure 4: Representative percent methylation by target
CUT&RUN-EM was performed as described in Figure 6. Global levels of DNA methylation associated with
each target (IgG, H3K4me3, and H3K36me3) are shown. The H3K36me3 positive control is highly
associated with DNA methylation at active gene bodies, while H3K4me3 has very low levels of DNA
methylation at active promoters.
Figure 5: Representative gene browser tracks
CUT&RUN-EM was performed as described in Figure 6. A 103 kb window is shown for IgG, H3K4me3, and
H3K36me3. CUT&RUN-EM produced the expected genomic distribution for each chromatin protein, whereby
H3K36me3 peaks are highly enriched for methylated DNA (red) in active gene bodies, and H3K4me3 peaks
are typically unmethylated DNA (blue) at active promoters. A whole genome EM-seq (WGEM) track
showing genome-wide DNA methylation is shown for reference. Images were generated using the
Integrative Genomics Viewer (IGV, Broad Institute).
Figure 6: CUT&RUN-EM methods
CUT&RUN-EM was performed using the CUTANA™ Multiomic CUT&RUN Controls Set and the CUTANA™
ChIC/CUT&RUN Kit (EpiCypher 14-1048) starting with 500k K562 cells and 0.5 µg of IgG (EpiCypher
13-0042), H3K4me3 (EpiCypher 13-0060), or H3K36me3 (EpiCypher 13-0058) antibodies. Library
preparation was performed with 1 ng of DNA using the NEBNext® Enzymatic Methyl-seq v2 Kit (NEB
E8015). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2x50 bp). Sample
sequencing depth was 31.5 million reads (IgG), 21.3 million reads (H3K4me3), and 20.6 million reads
(H3K36me3). Data were aligned to the hg38 genome using Bowtie2. Data were filtered to remove
duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions. Validation data are
representative where noted. Refer to the product page for 13-0058 for the lot-specific validation
approach for H3K36me3 antibody.
Set Contents
| Item | Cat. No. | QTY |
|---|---|---|
| CUTANA™ H3K36me3 Positive Control Antibody | 13-0058-03 | 8 rxn |
| CUTANA™ CpG Methylated pUC19 Fragmented Control DNA | 18-8001-05 | 20 µL |
| CUTANA™ CpG Unmethylated Lambda Fragmented Control DNA | 18-8002-05 | 20 µL |
| CUTANA™ 0.1X TE Buffer | 21-1025-05 | 2 x 2 mL |
Recommended Accessory Products
| Item | Cat. No. |
|---|---|
| CUTANA™ ChIC/CUT&RUN Kit | 14-1048/14-1048-24 |
| NEBNext® Enzymatic Methyl-seq v2 Kit | NEB E8015 |