Complete Step-by-Step Guide to CUT&RUN

Table of Contents
- What is CUT&RUN?
- What’s the difference between CUT&RUN and ChIP-seq?
- The 8 Basic Steps of CUT&RUN
- How To Design and Optimize CUT&RUN Assays
- Next Steps for Getting Started with CUT&RUN
What is CUT&RUN?
CUT&RUN, or Cleavage Under Targets and Release Using Nuclease, is a groundbreaking approach for ultra-sensitive genomic mapping of chromatin targets. CUT&RUN was developed in Dr. Steven Henikoff’s group1-3, and builds off immunotethering technology (chromatin immunocleavage, or ChIC) developed by Dr. Ulrich Laemmli4. In CUT&RUN, Protein A and Protein G are fused to micrococcal nuclease (pAG-MNase) and used to selectively cleave antibody-labeled chromatin in intact cells. The clipped fragments diffuse into solution, where they can be separated from cells, purified, and analyzed by next-generation sequencing (NGS).
CUT&RUN often draws comparisons to ChIP-seq since both assays identify the genomic location of chromatin-associated proteins (CAPs) and histone post-translational modifications (PTMs). However, CUT&RUN is fundamentally distinct from ChIP and provides a more robust approach for chromatin mapping.
What’s the difference between CUT&RUN and ChIP-seq?
- Lower cell requirements. CUTANA™ CUT&RUN assays generate high-resolution profiles using as few as 5,000 cells, and lower cell numbers have been reported in the literature5,6.
- Improved signal-to-noise. Separation of target-bound chromatin from bulk cells means lower background and robust signal – even when using fewer cells.
- Reduced cost. High signal-to-noise enables confident peak calling with ~10-fold fewer sequencing reads and less primary antibody.
- Less optimization. The entire assay is performed in intact cells (or nuclei) immobilized on a solid support. These advances eliminate the most technically challenging steps of ChIP-seq, including cell lysis, chromatin fragmentation, and immunoprecipitation (IP).
- Faster. CUT&RUN provides a user-friendly, streamlined protocol that allows scientists to go from cells to sequencing data in less than four days (vs. weeks for ChIP-seq). CUT&RUN can also be automated for greater throughput7,8.
CUT&RUN opens up entirely new avenues of research that were impractical using ChIP-seq. The low cell requirements make it feasible to analyze rare or precious samples (e.g. primary tissues, FACS-sorted cells) and to expand studies to additional targets. The increased throughput and reduced costs also support scaled epigenomics, which is key for chromatin mapping applications in clinical research and drug development. And of course, the improved signal and reduced background is a crucial advance that will drive novel applications of epigenomics datasets.
The 8 Basic Steps of CUT&RUN
Step 1: Immobilize cells. Cells are bound to magnetic beads coated with Concanavalin A (ConA), a lectin that binds to cell surface proteins. This supports high-throughput formatting and simplifies the separation of cells from on-target chromatin in Step 6.
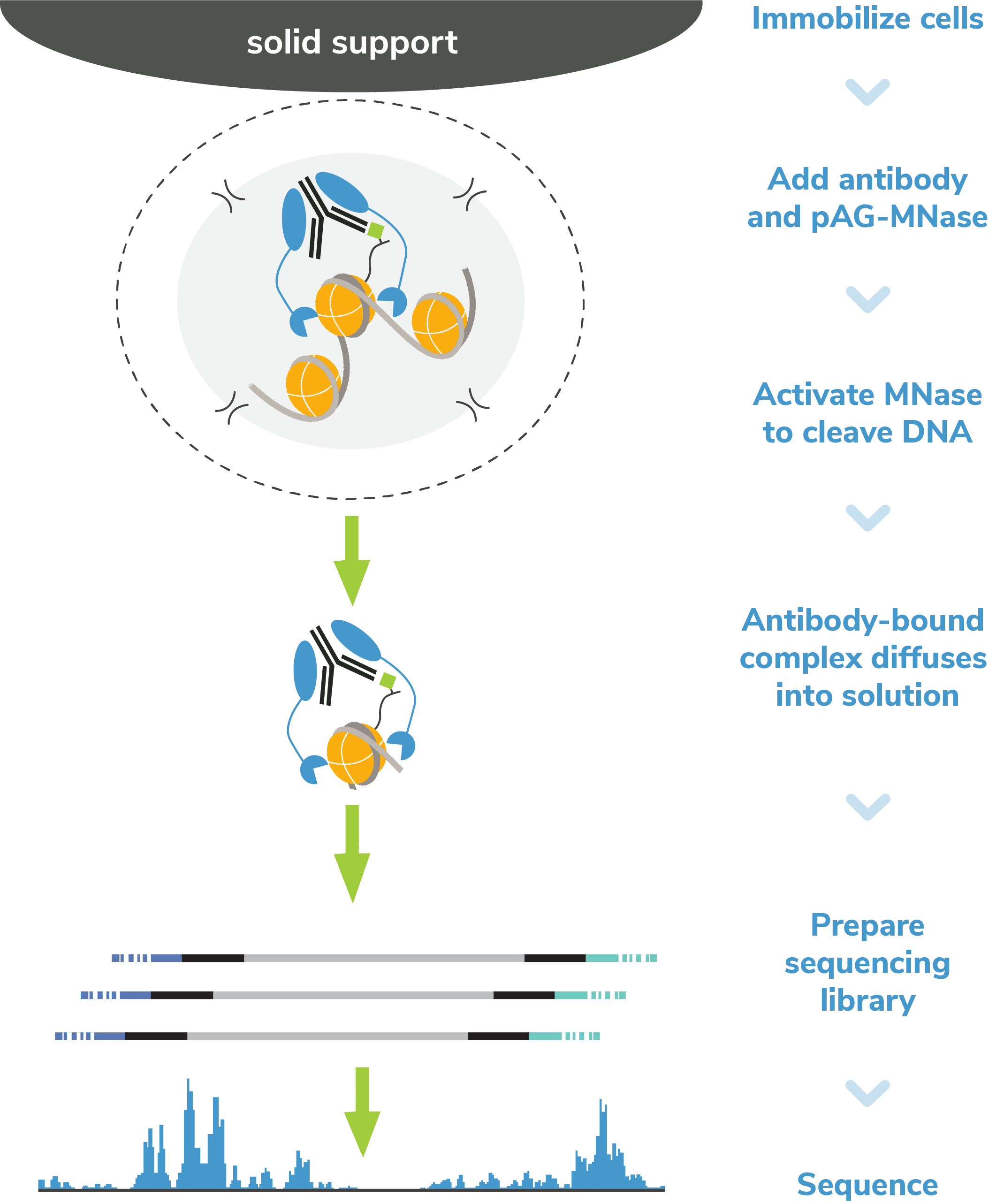
Step 2: Permeabilize cells. Immobilized cells are treated with a buffer containing digitonin, a nonionic detergent that permeabilizes cell membranes at low concentrations. The cell is often depicted with small holes in the lipid bilayer (Figure 1). This step allows antibody and pAG-MNase to bind chromatin during Steps 3 & 4 and MNase-digested DNA to diffuse into solution in Step 5.
CRITICAL STEP: Optimizing the digitonin concentration is key for successful CUT&RUN assays. Under-digestion prevents pAG-MNase from entering cells, while over-digestion may lyse cells. See our suggestions on assay optimization and quality control steps in the next section.
Step 3: Incubation with target-specific antibody. An antibody to the target of interest is added to the reaction and incubated overnight at 4˚C. We suggest including a negative control (e.g. IgG) and a positive control (e.g. H3K4me3, H3K27me3) in every experiment. Additional notes on antibody selection and assay controls are detailed in the optimization section of this blog.
CRITICAL STEP: The quality of antibody is crucial to CUT&RUN, since only antibody-bound chromatin is fragmented and collected for sequencing. Antibodies with low enrichment efficiency and/or specificity generate low yields and poor sequencing results.
Step 4: Add pAG-MNase. The following day, bead-bound cells are washed and then pAG-MNase is added to the reaction. The immunoglobulin binding properties of pAG act to “tether” MNase to antibody-bound chromatin. To prevent nonspecific cleavage, the cell/bead mixture is washed several times following pAG-MNase incubation.
Step 5: pAG-MNase Activation. Calcium (Ca2+) is added to the reaction to activate MNase, which cleaves DNA proximal to where the antibody is bound. Cleaved chromatin fragments diffuse into the supernatant, while remaining bulk chromatin remains inside the bead-immobilized cells.
Because MNase is a processive enzyme, the reaction must be quenched to prevent over-digestion of released DNA. Following pAG-MNase incubation, a Stop Buffer containing EDTA and EGTA is added to chelate free calcium ions and halt enzymatic activity.
Step 6: DNA purification. Isolation of CUT&RUN enriched DNA is straightforward since the cells remain bound to magnetic ConA beads. Bead-coupled cells containing bulk chromatin are magnetically separated from the clipped target DNA, which remains in solution. Target DNA is purified using a column clean up kit optimized for small fragments and quantified with a fluorometric assay (e.g. ThermoFisher Qubit™).
Important Note: Before library prep, DNA yields are the best indicator of assay success. In ChIP workflows, it is standard practice to determine fragment distribution of isolated ChIP DNA (via Agilent TapeStation® or Bioanalyzer®) and enrichment of known targets (via qPCR). However, these strategies are NOT robust indicators of CUT&RUN success and are NOT recommended. In fact, raw CUT&RUN DNA yields are usually below the sensitivity limit for TapeStation/Bioanalyzer analysis.
EpiCypher also checks that the yields from positive controls and experimental targets are greater than the IgG negative control reaction, even if only slightly higher. After measuring CUT&RUN DNA concentration, proceed to library prep.
Step 7: CUT&RUN library prep. Purified CUT&RUN DNA is repaired, ligated to sequencing adapters, and PCR-amplified to generate NGS libraries. PCR is performed using parameters optimized for low CUT&RUN yields and small fragment sizes, and barcoded primers are used to enable multiplexed sequencing. EpiCypher’s Library Prep Kit is specifically optimized for CUT&RUN and streamlines your workflow.
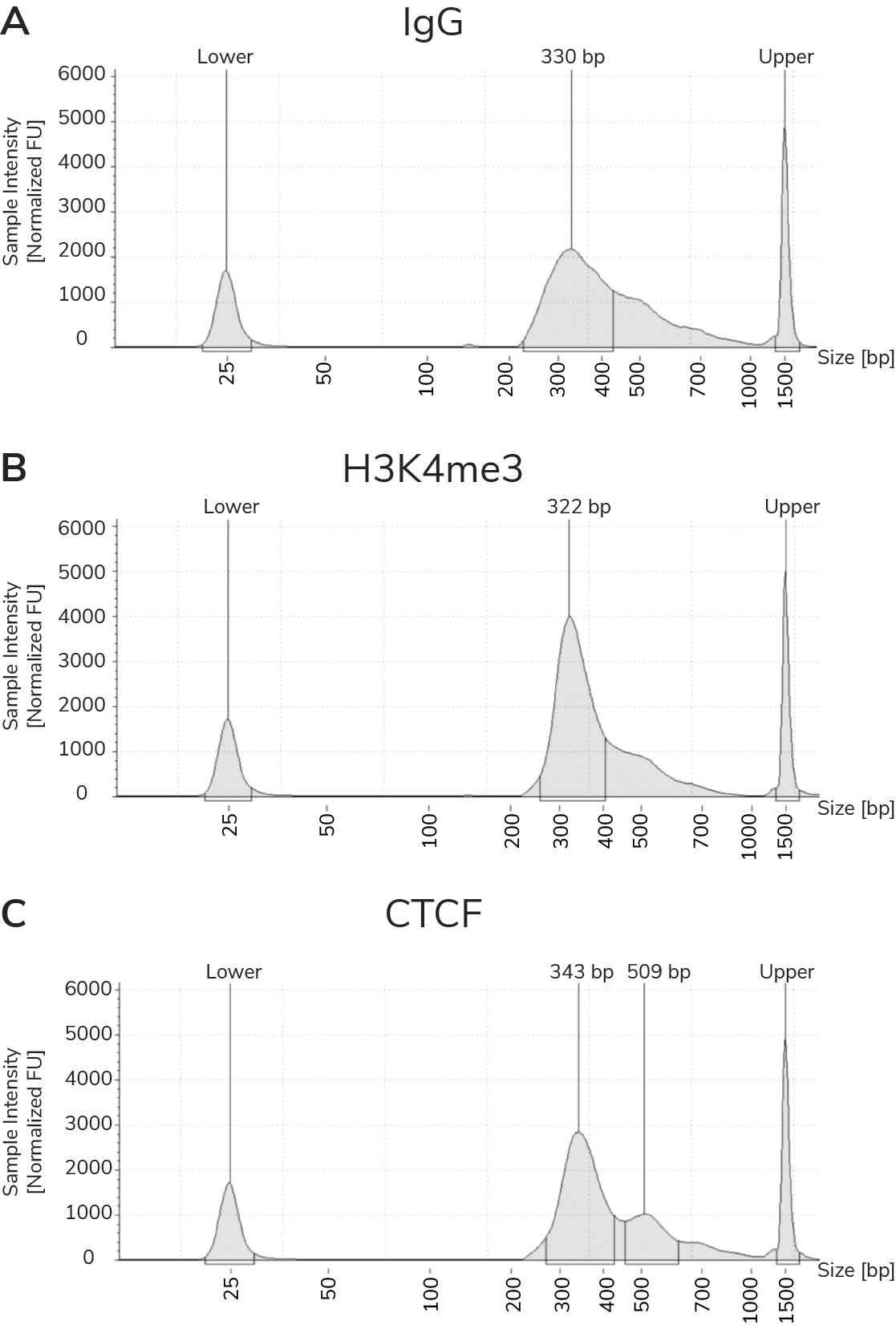
Prior to NGS, the best indicator of assay success is fragment size distribution of final prepared libraries. The fragment size distribution and concentration of final CUT&RUN libraries is confirmed using capillary electrophoresis (e.g. Agilent Bioanalyzer or TapeStation). Since MNase digests fragments to nucleosome-level resolution, the average peak size is typically ~300 bp (~170 bp fragmented DNA + adapters). Examples are provided in Figure 2; library prep is discussed in additional detail below.
Step 8: Next-generation sequencing (NGS). Libraries are pooled at equimolar ratios and loaded onto the desired platform for NGS. Only 3-8 million reads per sample are required for robust signal over background (vs. >20 million for ChIP-seq), allowing users to multiplex 10s-100s of samples in a single run.
Learn more about our CUT&RUN Kit
How to Design and Optimize CUT&RUN Assays
CUT&RUN is simple to optimize compared to ChIP-seq, largely due to the elimination of cross-linking, chromatin fragmentation, and immunoprecipitation. These three ChIP steps require difficult, time-consuming optimization, introduce unknown variability, and lead to high background in ChIP assays.
Bypassing these steps in CUT&RUN dramatically reduces background and increases sensitivity, enabling analysis of low cell numbers at reduced sequencing depths. It also makes CUT&RUN a faster, easier, and less expensive technique! EpiCypher has streamlined this strategy with our user-friendly CUTANA™ CUT&RUN protocols, kits, and reagents, making CUT&RUN profiling accessible to more researchers.
However, for robust CUT&RUN profiling, certain steps require optimization. We have broken these down into four main steps: sample preparation, assay controls, antibody selection, and DNA purification and analysis.
I. Sample Preparation
Number of Cells. We recommend starting with 500,000 native (unfixed) cells, particularly when mapping new targets or using new cell types. Following initial validation of workflows using 500,000 cells and control antibodies, cell numbers can be reduced following the recommendations in the CUT&RUN Kit manual and protocol.
Note on cell types: EpiCypher has developed several protocol variations to ensure reliable CUT&RUN data from diverse biological samples. We have recommendations for using adherent cells, tissues, immune cells, nuclei, and frozen cells/nuclei in Appendix II of our CUT&RUN Kit manual.
Notes on using tissues: CUTANA CUT&RUN Kits and protocols are compatible with tissues. The primary requirement when using tissues is to process samples into a monodispersion of cells. See Appendix II of the CUT&RUN Kit manual for further information.
Replicates. CUT&RUN is very robust and reliable – two biological replicates per target are usually sufficient (e.g. cells from two mutant mice).
Cross-linking. CUT&RUN is a native technique, meaning that it performs best on unfixed cells (or nuclei). This is a major advantage compared to ChIP, which typically requires heavy cross-linking to stabilize target associations with DNA. Cross-linking and chromatin fragmentation are major contributors to high background, low yields, artifacts, and/or data variability in ChIP. These steps aren’t required for CUT&RUN, which streamlines the assay, maximizes on-target DNA recovery, and allows for reduced cell numbers.
Cross-linking recommendations for CUT&RUN: There are some instances where light cross-linking can be useful in CUT&RUN, such as for stabilizing potentially labile PTMs (e.g. histone acetylation) and acetyl-binding proteins (e.g. bromodomains). Note that we always recommend trying native conditions first, or at least in parallel with cross-linked samples. See this blog and our cross-linking protocol for detailed information.
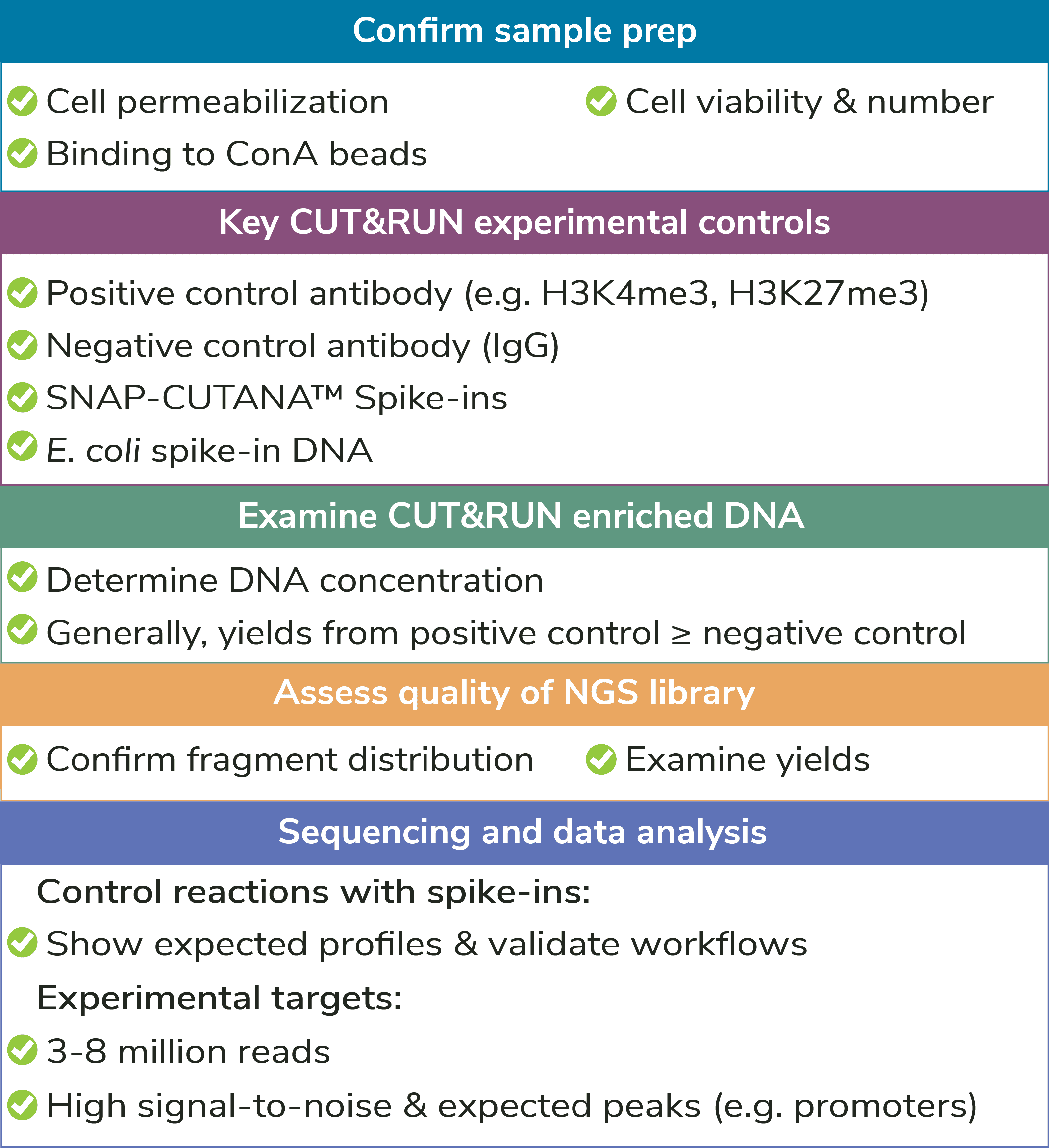
Optimization of Cell Permeabilization. This is a key step of the CUT&RUN protocol, as the cell membrane must be pervious to antibodies, pAG-MNase, and allow release of cleaved DNA, yet intact enough to prevent bulk chromatin dissociation. But don’t stress – EpiCypher provides a step-by-step protocol using a simple Trypan Blue staining procedure in the CUTANA™ CUT&RUN Kit Manual, along with detailed examples. For a full list of recommended quality control checks, see Figure 4.
II. Controls
All CUT&RUN experiments should be performed with appropriate controls to ensure the assay performs as expected. Note: All of these controls are included with the CUTANA™ CUT&RUN Kit!
Positive and Negative Control Reactions. Reactions using negative control (IgG) and positive control (H3K4me3 or H3K27me3) antibodies should be included in every experiment to validate workflows. EpiCypher also recommends adding the SNAP-CUTANA™ K-MetStat Panel of spike-in controls to these positive and negative control reactions.
What is the SNAP-CUTANA™ K-MetStat Panel? The K-MetStat Panel is a pool of 16 modified recombinant nucleosomes carrying widely studied methyl-lysine histone PTMs. Each nucleosome is wrapped with DNA containing a PTM-specific barcode to enable detection by NGS. Importantly, the K-MetStat panel is conjugated to paramagnetic beads, which allows it to be processed alongside immobilized cells in CUT&RUN. The panel is added to CUT&RUN control reactions just prior to addition of primary antibody (Figure 3).
The spike-ins replicate the physiological targets of CUT&RUN assays (nucleosomes) and carry both on- and off-target epitopes for H3K4me3 and H3K27me3 control reactions. Thus, the spike-in results provide a quantitative readout of PTM recovery, and can be used to directly validate CUT&RUN workflows. Poor results from K-MetStat Spike-ins can guide troubleshooting experiments. See the SNAP-CUTANA™ Spike-in User Guide for more information.
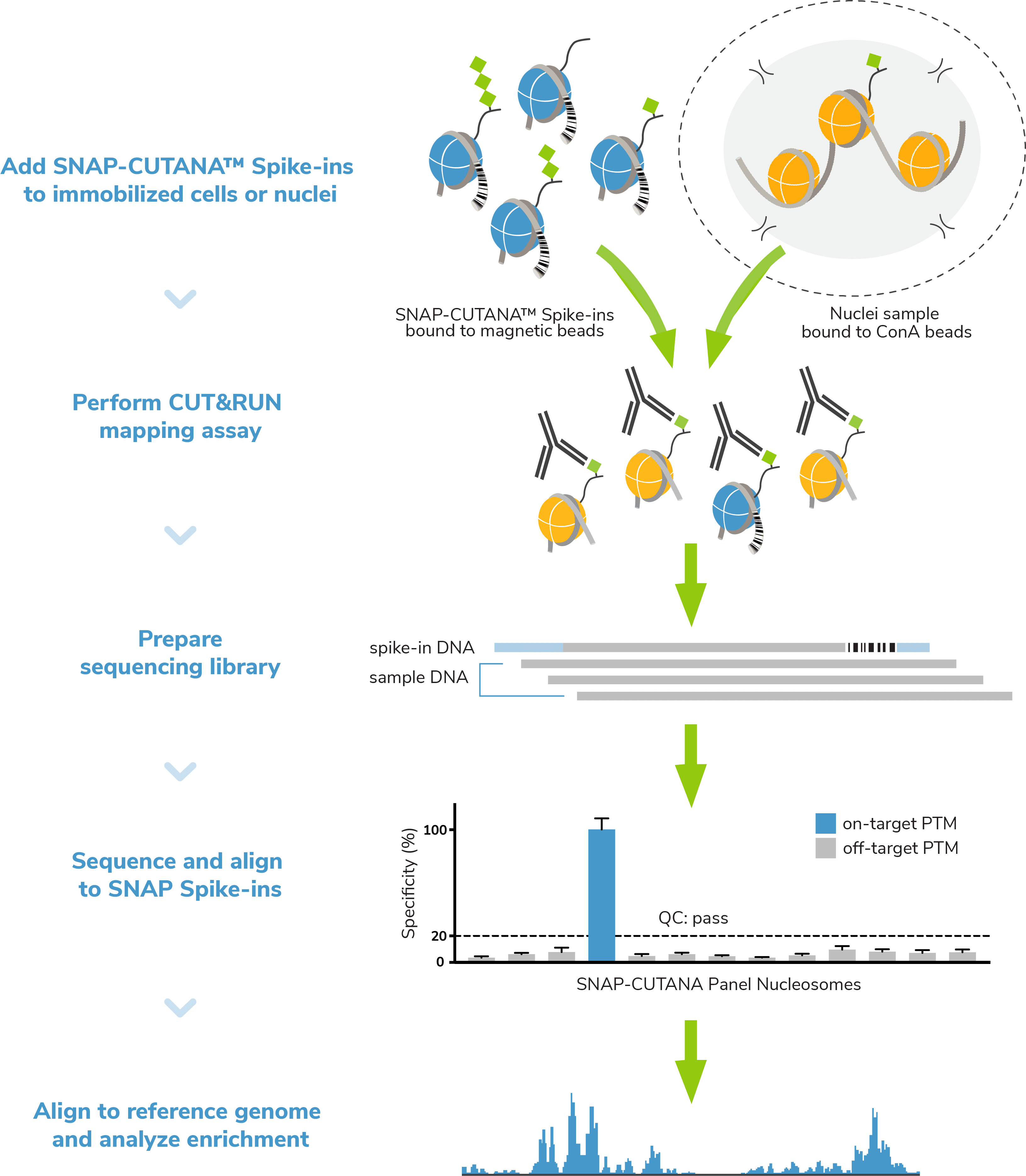
Spike-in controls. Spike-in controls are essential for all genomics assays. The CUTANA CUT&RUN Kit includes includes E. coli spike-in DNA, which can be added as a control for library prep and aid in NGS normalization. The K-MetStat panel can also be added to experimental reactions profiling histone methyl-lysine PTMs to examine antibody specificity, signal over background, and assay variability (Figure 3).
Quality Control Checks: EpiCypher has incorporated multiple quality control checks to help ensure assay success (Figure 4). For instance, we have developed a simple Trypan Blue staining protocol to confirm sample binding to ConA beads – an important step of CUT&RUN workflows. A full list of quality control checks, both before AND after sequencing, can be found in the CUT&RUN Kit manual and protocol.
III. Antibody Selection
Select a target-specific antibody. As with ChIP-seq, a quality antibody is essential for generating robust and reliable profiles. We have covered principles for antibody selection extensively in the CUT&RUN protocol, kit manual, and previous blogs. Briefly:
- Performance in other assays, such as ChIP, does NOT guarantee success in CUT&RUN! The antibody should demonstrate high-specificity (low cross-reactivity) and high efficiency (comprehensive target recognition) in CUT&RUN.
- EpiCypher offers CUTANA™ CUT&RUN antibodies against histone PTMs and multiple target classes of chromatin-associated proteins (Figure 2). These antibodies have been extensively screened for robust performance in CUT&RUN assays. Contact us for additional antibody recommendations not found in our current catalog.
- When analyzing a new target in the absence of validated antibodies, we recommend sourcing 3-5 antibodies from various reputable vendors that bind unique epitopes. Test these antibodies in parallel CUT&RUN assays with cells known to express the target protein. Then, select the best antibody based on overall yield, signal over background, and peak structure/enrichment.
Shop CUTANA™ CUT&RUN Antibodies
IV. DNA Isolation and Analysis
DNA purification. Yields from CUT&RUN are much lower compared to ChIP, and DNA fragments can also be much smaller, particularly for transcription factors. In these cases, it is important to use a DNA purification kit that is optimized for low DNA concentrations and small fragment sizes. EpiCypher’s CUTANA CUT&RUN Kit includes DNA purification columns and buffers that are specifically designed to retain fragments down to 50 bp. The protocol also elutes in low volume, which maximizes DNA concentration for subsequent library prep.
Note: We also offer these DNA purification reagents in a separate CUTANA™ DNA Purification Kit!
Library prep. Library prep is a standard part of genomics assays, and there are numerous kits and multiplexing strategies available. However, the low yields and small fragments from CUT&RUN make it difficult to utilize existing library prep kits. For instance, it isn’t clear if adapter concentrations should be adjusted for low DNA inputs, or the optimal SPRI bead ratio to use for enriching small CUT&RUN fragments while avoiding adapter-dimer contaminants. To address these concerns, EpiCypher has launched all-inclusive Library Prep Kits specifically optimized for CUTANA CUT&RUN assays. You can read more about library kit development in another blog post.
Sequencing Depth. The number of sequencing reads depends on several factors, including the number of cells, target abundance, and antibody quality. For most targets, 3-8 million paired-end reads are sufficient and will allow you to multiplex more samples per run. If you have further questions about sequencing depths or CUT&RUN library prep, see the FAQs section of the CUT&RUN Library Prep Kit Manual.
Next Steps for Getting Starting with CUT&RUN
CUT&RUN generates high-quality data with fewer steps, cells, and sequencing reads with less optimization compared to ChIP-seq – which means you can profile more targets, sequence more samples, and save valuable time and resources. Despite its recent development, CUT&RUN has been successfully used by EpiCypher and many others to map histone PTMs, histone variants, and chromatin-associated proteins.
CUT&RUN also has a sister technology, CUT&Tag, which is even more streamlined and ideal for specific applications, including single-cell profiling. We will describe the CUT&Tag workflow in the next blog!
Here are some additional resources to learn more and get started with CUT&RUN:
- CUTANA™ CUT&RUN Products
- CUT&RUN Protocols and Manuals
- EpiCypher Blog Posts
Ready to get started with CUT&RUN?
Click here to request more information about EpiCypher’s CUTANA CUT&RUN Assays and/or any other products and services:
References
- Skene, P. J., and Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017). PubMed PMID: 28079019.
- Skene, P.J. et al. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006-1019 (2018). PubMed PMID: 29651053.
- Meers M.P. et al. Improved CUT&RUN chromatin profiling tools. eLife 8, e46314 (2019). PubMed PMID: 31232687.
- Schmid, M. et al. ChIC and ChEC: Genomic Mapping of Chromatin Proteins. Mol. Cell 16, 147–157 (2004). PubMed PMID: 15469830.
- Hainer, S. J. et al. Profiling of Pluripotency Factors in Single Cells and Early Embryos. Cell 177, 1319-1329.e11 (2019). PubMed PMID: 30955888.
- Patty, B. J., and Hainer, S. J. Transcription factor chromatin profiling genome-wide using uliCUT&RUN in single cells and individual blastocysts. Nat. Protoc. 16, 2633–2666 (2021). PubMed PMID: 33911257.
- Janssens, D. H. et al. Automated CUT&Tag profiling of chromatin heterogeneity in mixed-lineage leukemia. Nat. Genet. 4, 1586-1969 (2021). PubMed PMID: 34663924.
- Janssens, D. H. et al. Automated in situ chromatin profiling efficiently resolves cell types and gene regulatory programs. Epigenetics and Chromatin 11, 74 (2018). PubMed PMID: 30577869.
