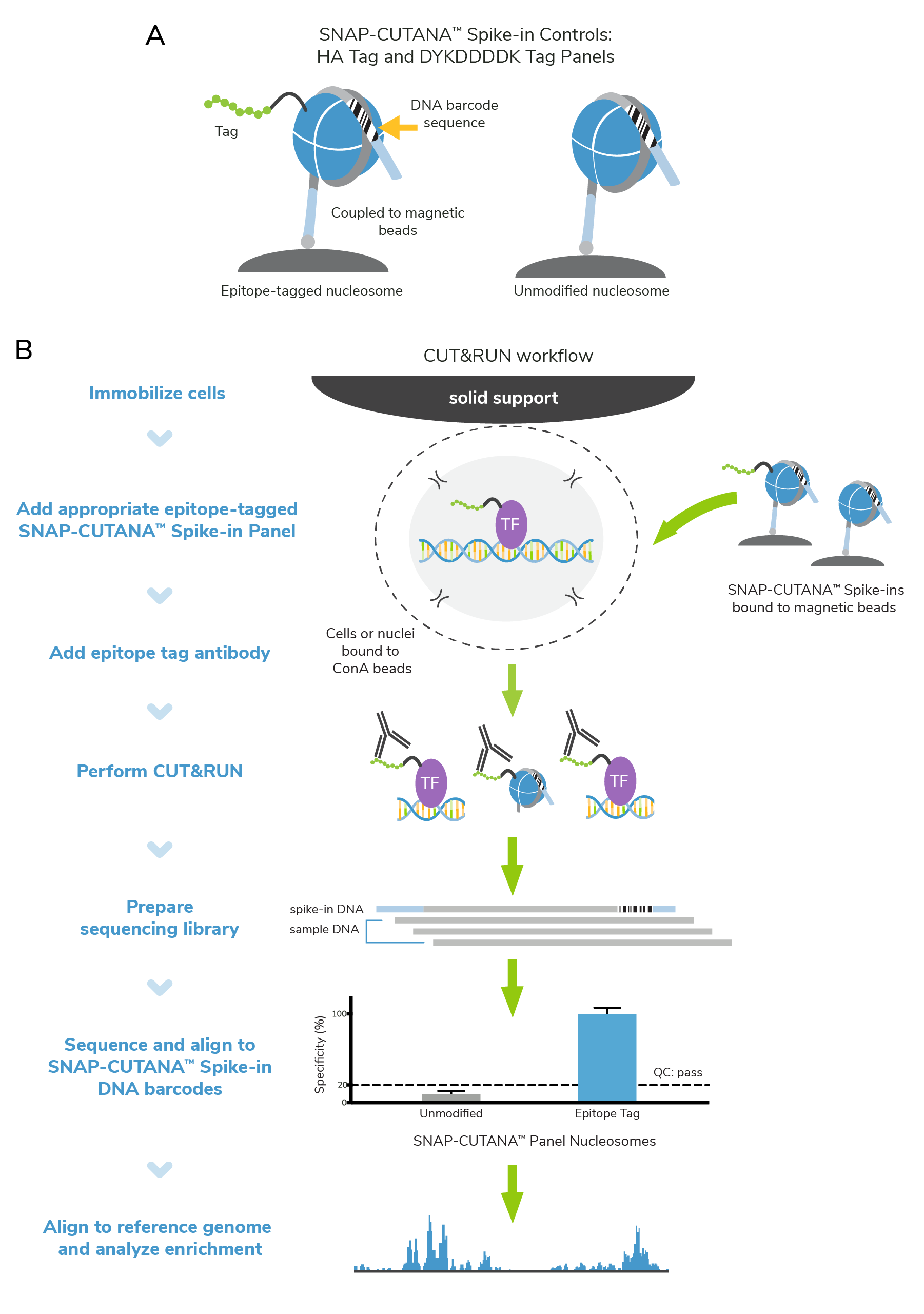
Improved controls for CUT&RUN profiling of epitope-tagged proteins

Protein tags have multiple uses and advantages in molecular biology1,2. They can improve protein stability in vitro, streamline purification processes, and allow interrogation of protein-protein interactions. Tags are particularly useful for detecting proteins that lack reliable and specific antibodies. In epigenomics research, HA and DYKDDDDK (FLAG®) epitope tags are frequently used to study the genome-wide enrichment of transcription factors, modifying complexes, or other chromatin-associated proteins. The integration of defined controls is a critical part of optimizing and troubleshooting workflows. Here, we share how our new epitope-tagged SNAP-CUTANA™ Spike-in Controls helped our scientists troubleshoot a recent CUT&RUN experiment.
Shop new HA Tag spike-in controls
How do epitope tags work?
An expression vector encoding the epitope-tagged protein is expressed stably or transiently in the cells of interest. Importantly, epitope tags can only be used to detect recombinant proteins — endogenous proteins do not carry epitopes.
Epitope-tagged proteins are characterized using antibodies that recognize the protein tag. Expression is confirmed by immunoblot, while cellular localization can be further validated by immunofluorescence or immunohistochemistry. Functional analysis often requires immunoprecipitation experiments, in which the tag antibody is used to pull down the epitope-tagged protein, followed by immunoblot for known interacting partners.
In the study of chromatin-associated proteins, however, more thorough validation is often required. Read on to learn more!
Why are controls important when mapping epitope-tagged proteins?
Despite the many advantages of epitope tags, their application comes with several caveats. Tags can impact protein folding, cellular localization, and other regulatory mechanisms1,2. Regulatory elements used to express integrated constructs can become silenced over many passages, a phenomenon that also challenges cell and gene therapy research3. It is crucial to include experimental controls that can confirm tagged protein function, antibody binding activity, and monitor CUT&RUN protocol steps.
To this end, EpiCypher developed epitope-tagged SNAP-CUTANA™ Spike-In Control Panels, which uses our proprietary nucleosome spike-in technology to monitor epitope antibody performance and CUT&RUN assay success. These panels contain epitope-tagged and unmodified nucleosomes coupled to magnetic beads (Figure 1), streamlining their integration in CUT&RUN workflows. Recovery of epitope-tagged and unmodified nucleosomes is determined via their unique DNA barcode sequences. Spike-in results will help you understand whether your CUT&RUN experiment was successful, and your antibody enriched for the target epitope. The process is straightforward:
- SNAP-CUTANA™ Spike-ins are added to immobilized cells, just prior to adding epitope antibody. No other protocol modifications are required.
- Epitope antibodies bind their target in cells and in the spike-in panel, enabling targeted cleavage and release by pAG-MNase.
- Enrichment of epitope-tagged vs. unmodified spike-in nucleosomes is determined from CUT&RUN sequencing data via analysis of DNA barcode sequences.
Behind-the-scenes: SNAP-CUTANA™ HA Tag Spike-ins help troubleshoot CUT&RUN experiments
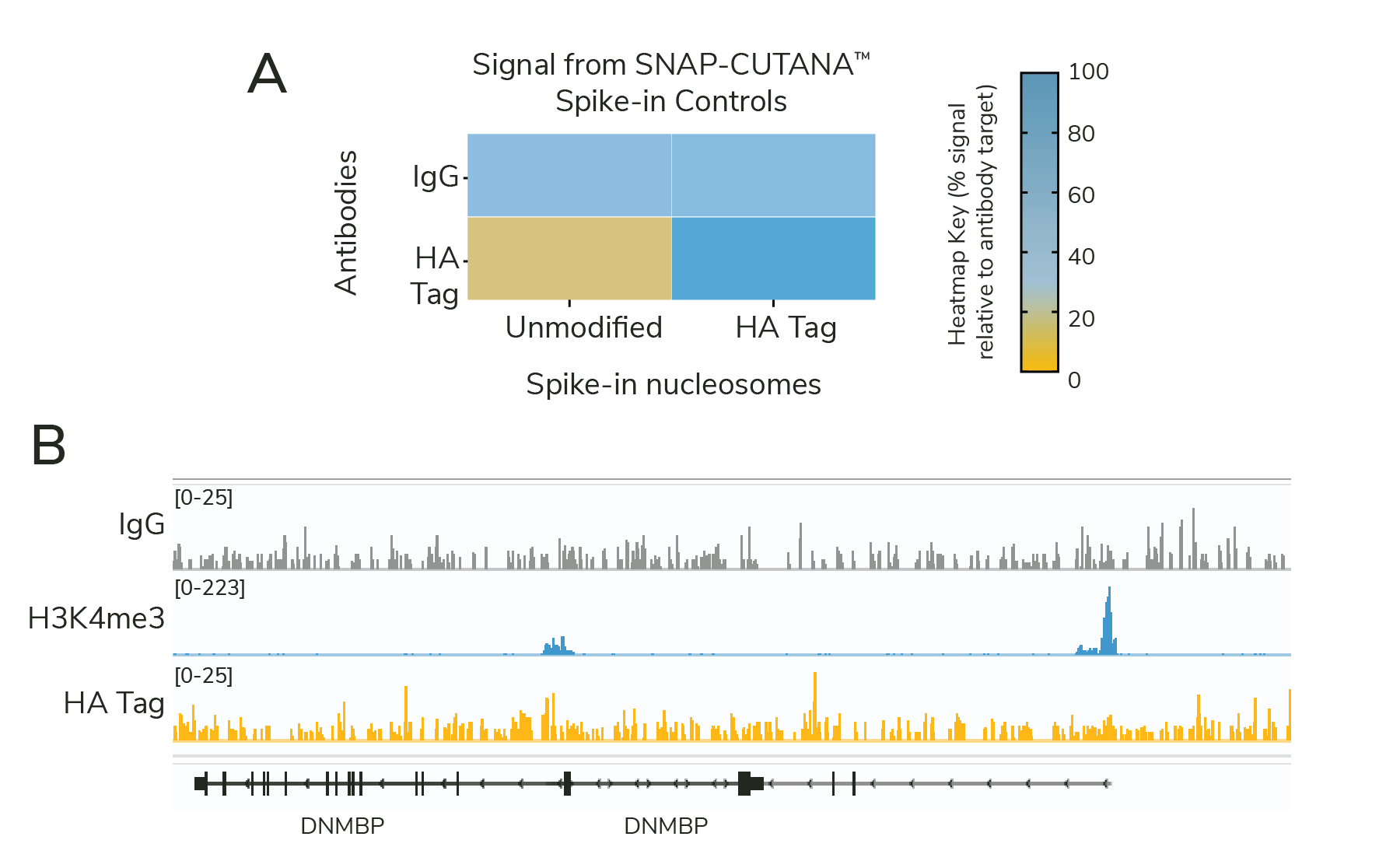
SNAP-CUTANA™ Spike-ins can help pinpoint problems with failed CUT&RUN assays. This is nicely illustrated in experiments using our CUT&RUN-validated HA Tag antibody (EpiCypher 13-2010) and corresponding SNAP-CUTANA™ HA Tag Panel (EpiCypher 19-5002).
For this experiment, EpiCypher scientists used MDA-MB-231 breast cancer cells expressing HA-tagged GATA3 (HA-GATA3 cells, kindly provided by Motoki Takaku, PhD). These cells have been independently validated and published, offering a robust system for HA antibody validation4.
We set up CUT&RUN reactions using HA-GATA3 cells and HA Tag and IgG antibodies, each spiked with the SNAP-CUTANA™ HA Tag Panel. H3K4me3 antibody (EpiCypher 13-0060) was used as a positive control, as it should produce robust CUT&RUN profiles regardless of HA-GATA3 expression.
Surprisingly, analysis of HA sequencing data revealed poor signal and minimal peaks (Figure 2B; n=18 peaks using MACS2). What could be wrong? Our scientists examined each assay control to narrow down potential issues:
- The HA Tag antibody specifically enriched for corresponding HA spike-in nucleosomes (Figure 2A), confirming multiple aspects of the CUT&RUN workflow: buffer prep, antibody binding, pAG-MNase activity, and downstream library prep steps.
- IgG antibodies showed no preference among nucleosomes in the panel and generated minimal background in sequencing tracks (Figure 2A & 2B), further validating our CUT&RUN technique.
- The H3K4me3 positive control antibody generated sharp peaks at active transcription start sites, as expected (Figure 2B). This control, combined with the low background from IgG, suggests that samples were correctly prepared, with minimal cell lysis and/or loss during the protocol.
Importantly, all controls provided confidence in our CUT&RUN approach. This led us to one main hypothesis – an issue with HA-GATA3 expression. As noted above, transgenes can become silenced over time due to a variety of mechanisms, or have issues with protein folding, localization, and/or stability. In addition, the HA-GATA3 construct uses the CMV promoter, which can be impacted by glucose levels in cell culture media5.
To address this issue, our scientists took multiple approaches:
- Obtained fresh vials of HA-GATA3 cells, to avoid potential issues with transgene expression over prolonged passaging.
- Sourced MDA-MB-231 cells transduced with empty vector as an additional control.
- Cultured cells in appropriate media, per published guidelines.
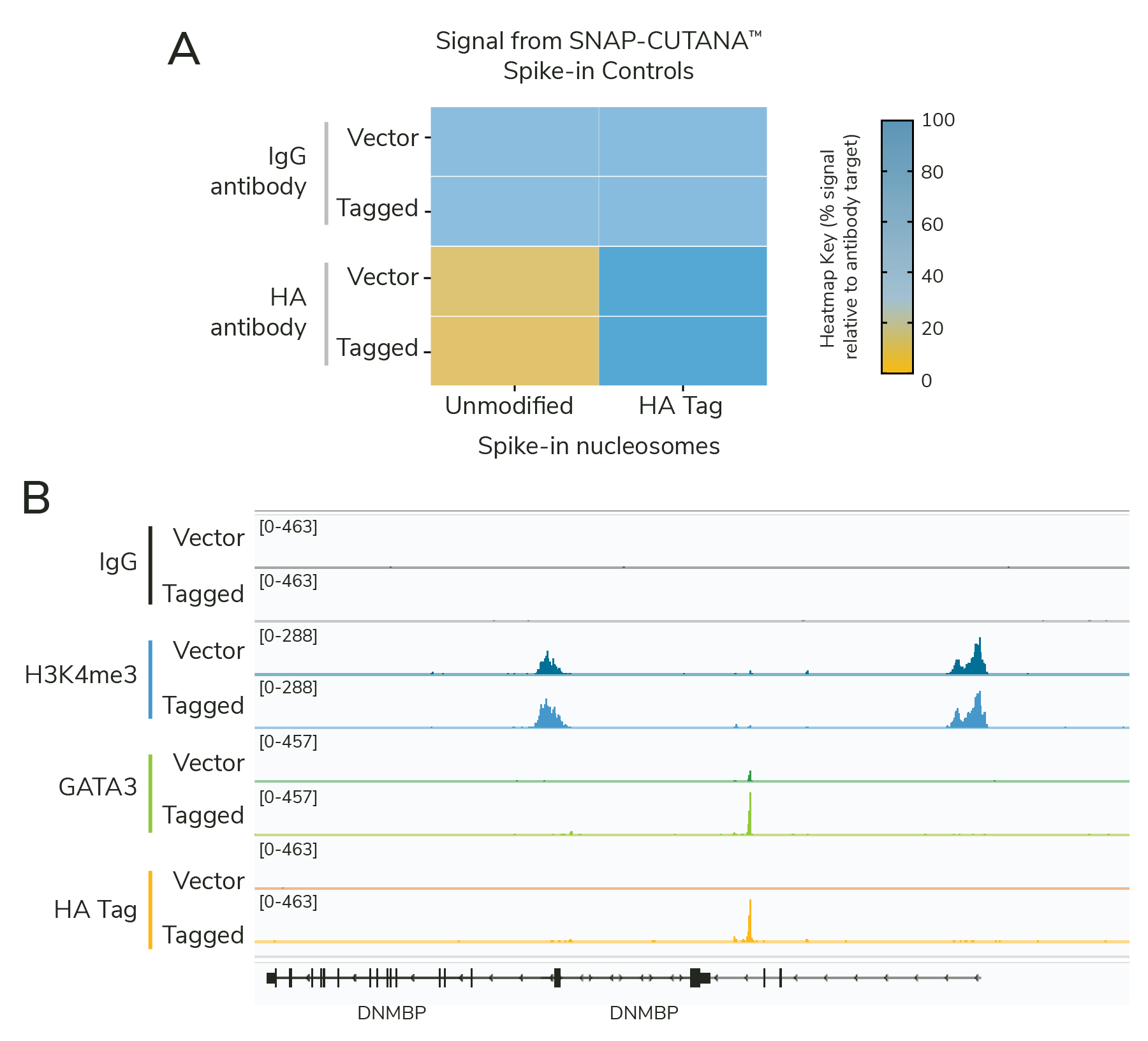
- Added GATA3 antibodies in CUT&RUN experiments to confirm expression and enrichment of HA-tagged GATA3. Note that the MDA-MB-231 cells have very low levels of endogenous GATA3, so nearly all signal is derived from the tagged construct.
- Included all CUT&RUN controls from the previous experiment: HA Tag and IgG antibodies spiked with the HA Tag Panel, and H3K4me3 positive control antibody spiked with the K-MetStat Panel.
The results showed that the cell line optimization was successful. HA-GATA3 cells generated highly similar profiles using HA Tag and GATA3 antibodies, together demonstrating robust expression of the GATA3 construct (Figure 3B). Spike-in results validate the specific enrichment of HA-tagged vs. unmodified proteins and confirm workflow success (Figure 3A). Empty vector cells had little to no GATA3 or HA Tag signal, as expected, but showed robust H3K4me3 enrichment, confirming sample prep and CUT&RUN technique.
As noted above, our CUT&RUN assay had always worked. The SNAP-CUTANA™ Spike-ins were critical to determine what parts of the first experiment were successful, and narrow down potential areas for troubleshooting. Although it is difficult to pin down the precise reason our assays worked in this second CUT&RUN experiment, optimizing cellular conditions played a pivotal role.
Why are controls important when mapping epitope-tagged proteins?
In any scientific experiment, multiple things can go wrong – no one is perfect! Our SNAP-CUTANA Spike-in Controls are designed to help you flag problems with CUT&RUN reactions. Spike-in results narrow down potential causes and guide troubleshooting approaches, leading to better chromatin profiling data without the tediousness of testing every protocol step.
SNAP-CUTANA Spike-ins were originally developed for histone post-translational modifications (PTMs), such as the K-MetStat Panel, a physiological control for histone lysine methylation PTMs. Our expanded collection includes SNAP-CUTANA Spike-ins for DYKDDDDK (FLAG®) and HA tags, which help address the unique issues that arise when mapping epitope-tagged proteins.
We provide additional guidance in the SNAP-CUTANA Spike-in User Guide. Check out these resources to get started:
SNAP-CUTANA™ Spike-in Controls:
CUT&RUN Validated Antibodies:
SNAP-Certified™ H3K4me3 antibody
SNAP-Certified™ H3K27me3 antibody
Resources:
SNAP-CUTANA™ Spike in User Guide
CUTANA™ CUT&RUN Library Prep Kit
FLAG is a registered trademark of Merck KGaA, Darmstadt, Germany and ANTI-FLAG is a trademark of Sigma-Aldrich Co. LLC
References:
- Esposito D, Chatterjee DK. Enhancement of soluble protein expression through the use of fusion tags. Curr Opin Biotechnol 17, 353-8 (2006). https://doi.org/10.1016/j.copbio.2006.06.003
- Young CL et al. Recombinant protein expression and purification: a comprehensive review of affinity tags and microbial applications. Biotechnol J 7, 620-34 (2012). https://doi.org/10.1002/biot.201100155
- Cabrera A et al. The sound of silence: Transgene silencing in mammalian cell engineering. Cell Syst 13, 950-73 (2022). https://doi.org/10.1016/j.cels.2022.11.005
- Takaku M et al. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol 17, 36 (2016). https://doi.org/10.1186/s13059-016-0897-0
- Kimura M et al. Enhancive effects of D-glucose and its analogs on expression of d-glucose-unrelated transgenes in mammalian cells. J Biosci Bioeng 112, 194-201 (2011). https://doi.org/10.1016/j.jbiosc.2011.04.010
