![Heterotypic [H3 • H3K4me3,K14ac] Recombinant Nucleosome, Biotinylated](https://cdn11.bigcommerce.com/s-y9o92/images/stencil/500x500/products/1166/1238/Heterotypic_Nuc_white__67835.1723235454.png?c=2)

Heterotypic [H3 • H3K4me3] Recombinant Nucleosome, Biotinylated
{"url":"https://www.epicypher.com/products/nucleosomes/modified-designer-nucleosomes-dnucs/heterotypic-h3-h3k4me3-recombinant-nucleosome-biotinylated","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"id":1164,"bulk_discount_rates":[],"can_purchase":true,"meta_description":"Heterotypic nucleosomes, also referred to as “asymmetric nucleosomes,” contain sister histones with distinct histone variants and/or post-translational modifications (PTMs). Heterotypic nucleosomes represent an additional layer of the histone code, acting as substrates for multivalent reader proteins, participating in PTM crosstalk mechanisms, and influencing reader protein binding affinity through varying local target concentration. Recombinant heterotypic nucleosomes are useful for studying chromatin dynamics and transcriptional regulation. Heterotypic [H3 • H3K4me3] Recombinant Nucleosome, Biotinylated is a fully defined semi-synthetic nucleosome containing one unmodified histone H3 and one histone H3 with trimethylated lysine at position four. Heterotypic [H3 • H3K4me3] nucleosomes fail to recruit known H3K4me3 binding proteins, including members of the TFIID, SETD1, and SIN3A/B complexes; chromatin remodelers NURF, CHD1, and CHD8; and PHD finger protein PHF2, suggesting that heterotypic nucleosomes may regulate transcription by affecting the recruitment of various chromatin regulatory complexes. ","category":["Nucleosomes","Nucleosomes/Modified Designer Nucleosomes (dNucs™)"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1164/1236/Heterotypic_Nuc_white__46729.1723235178.png?c=2","alt":"Heterotypic [H3 • H3K4me3] Recombinant Nucleosome, Biotinylated"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1164","shipping":{"calculated":true},"num_reviews":0,"weight":"0.00 LBS","custom_fields":[{"id":"1329","name":"Pack Size","value":"25 µg"}],"sku":"16-0408","description":"<div class=\"product-general-info\">\n <ul class=\"product-general-info__list-left\">\n <li class=\"product-general-info__list-item\">\n <strong>Species: </strong>Human\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Source: </strong><i>E. coli</i> & synthetic DNA\n </li>\n </ul>\n <ul class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <strong>Tag: </strong>Biotinylated\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Molecular Weight: </strong>199,689 Da\n </li>\n </ul>\n</div>\n<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <p>\n Heterotypic nucleosomes, also referred to as “asymmetric nucleosomes,” contain sister histones with distinct\n histone variants and/or post-translational modifications (PTMs). In homotypic nucleosomes, or “symmetric\n nucleosomes,” each pair of sister histones bears the same PTM, set of PTMs, or histone variant.\n Histone-modifying enzymes, chromatin remodelers, and histone chaperones differentially modify sister\n histones or exchange unique histone variants to form heterotypic nucleosomes. Heterotypic nucleosomes have\n been found at promoters of developmental genes in undifferentiated embryonic stem cells and transcription\n start sites (TSSs) of approximately half of the genes in budding yeast [1]. Heterotypic nucleosomes\n represent an additional layer of the histone code, acting as substrates for multivalent reader proteins,\n participating in PTM crosstalk mechanisms, and influencing reader protein binding affinity through varying\n local target concentration. Recombinant heterotypic nucleosomes are useful for studying chromatin dynamics\n and transcriptional regulation.\n </p>\n <p>\n Heterotypic [H3 • H3K4me3] Recombinant Nucleosome, Biotinylated is a fully defined semi-synthetic nucleosome\n containing one unmodified histone H3 and one histone H3 with trimethylated lysine at position four. [H3 •\n H3K4me3] nucleosome consists of 147 base pairs of 601 sequence DNA [2] wrapped around an octamer of core\n histone proteins (two each of H2A, H2B, H3.2, and H4) to form a nucleosome, the basic repeating unit of\n chromatin. The DNA contains a 5’ biotin-TEG group, and histone H3.2 has a Cys to Ala substitution at\n position 110. Heterotypic [H3 • H3K4me3] nucleosomes fail to recruit known H3K4me3 binding proteins,\n including members of the TFIID, SETD1, and SIN3A/B complexes; chromatin remodelers NURF, CHD1, and CHD8; and\n PHD finger protein PHF2, suggesting that heterotypic nucleosomes may regulate transcription by affecting the\n recruitment of various chromatin regulatory complexes [3].\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/nucleosomes/16-0408-protein-gel.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img alt=\"16-0408-protein-gel\"\n src=\"/content/images/products/nucleosomes/16-0408-protein-gel.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 1: Protein gel data</strong><br />\n Coomassie stained SDS-PAGE gel of proteins in heterotypic [H3 • H3K4me3] nucleosome (1 µg)\n demonstrates the purity of histones in the preparation. Sizes of molecular weight markers and\n positions of the core histones (H2A, H2B, H3 • H3K4me3, and H4) are indicated.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/nucleosomes/16-0408-dna-gel.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img alt=\"16-0408-dna-gel\"\n src=\"/content/images/products/nucleosomes/16-0408-dna-gel.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 2: DNA gel data </strong><br />\n Heterotypic [H3 • H3K4me3] nucleosome resolved via native PAGE and stained with ethidium bromide to\n visualize DNA. Both lanes are from the same gel. <b>Lane 1</b>: Free DNA (EpiCypher <a\n href=\"/products/nucleosomes/nucleosome-assembly-601-sequence-dna-biotinylated\"\n target=\"_blank\">18-0005</a>; 75 ng). Free\n DNA is over 95% pure by densitometry. <b>Lane 2</b>: Intact heterotypic [H3 • H3K4me3] nucleosomes\n (400\n ng).\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/nucleosomes/16-0408-luminex-multiplexed.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\">\n <img alt=\"16-0408-luminex-multiplexed\"\n src=\"/content/images/products/nucleosomes/16-0408-luminex-multiplexed.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span>\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 3: Luminex multiplexed specificity profiling</strong><br />\n Heterotypic [H3 • H3K4me3] nucleosome was assessed using a Luminex® based approach. The panel\n comprises biotinylated designer nucleosomes individually coupled to color-coded Luminex MagPlex®\n beads. Histone antibody (EMD Millipore MAB3422), DNA antibody (EMD Millipore MAB030), and H3K4me3\n antibody (EpiCypher <a\n href=\"/products/antibodies/h3k4me3-antibody-snap-certified-for-cut-run-and-cut-tag\"\n target=\"_blank\">13-0060</a>) were added (x-axis), and a second layer of anti-IgG*Phycoerythrin\n (PE; BioLegend 406421 for anti-rabbit IgG or BioLegend 405307 for anti-mouse IgG) was used for\n detection. Data were generated using a Luminex FlexMAP 3D®. Histone and DNA antibodies bind\n nucleosomes in the panel equivalently, as expected. H3K4me3 antibody binds heterotypic [H3 •\n H3K4me3] nucleosomes at reduced strength in comparison to homotypic H3K4me3 nucleosomes (EpiCypher\n <a href=\"/products/nucleosomes/nucleosome-recombinant-human-h3k4me3-dnuc-biotinylated\"\n target=\"_blank\">16-0316</a>).\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img alt=\"16-0408-protein-gel\" src=\"/content/images/products/nucleosomes/16-0408-protein-gel.jpeg\"\n width=\"200\" class=\"image-picker__side-image image-picker__side-image_active\" role=\"button\" />\n <img alt=\"16-0408-dna-gel\" src=\"/content/images/products/nucleosomes/16-0408-dna-gel.jpeg\"\n class=\"image-picker__side-image\" role=\"button\" />\n <img alt=\"16-0408-luminex-multiplexed\"\n src=\"/content/images/products/nucleosomes/16-0408-luminex-multiplexed.jpeg\"\n class=\"image-picker__side-image\" role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Stable for six months at -80°C from date of receipt. For best\n results, aliquot and avoid freeze/thaws.\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Formulation</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 10 mM Tris-HCl pH 7.5, 25 mM NaCl, 1 mM EDTA, 2 mM DTT, 20%\n glycerol (13.6 µg protein, 25 µg DNA + protein).\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Application Notes</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <p>\n Heterotypic [H3 • H3K4me3] nucleosome is highly purified and suitable for a variety of applications,\n including use as a substrate in enzyme assays, high-throughput screening and inhibitor testing, chromatin\n binding studies, protein-protein interaction assays, structural studies, and in effector protein binding\n experiments. Trace amounts of TEV protease may be present (less than 5% of the preparation by densitometry,\n <b>Figure 1</b>). For a corresponding homotypic control, we recommend EpiCypher <a\n href=\"/products/nucleosomes/nucleosome-recombinant-human-h3k4me3-dnuc-biotinylated\"\n target=\"_blank\">16-0316</a>.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Gene & Protein Information</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>UniProt ID</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n H2A - P04908 (alt. names: H2A type 1-B/E, H2A.2, H2A/a, H2A/m)<br />\n H2B - O60814 (alt. names: H2B K, HIRA-interacting protein 1)<br />\n H3.2 - Q71DI3<br />\n H4 - P62805\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">References</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <strong>Background References:</strong>\n <br />\n [1] Valsakumar & Voigt. <em>Biochem. Soc. Trans.</em> (2024). PMID:\n <a href=\"https://pubmed.ncbi.nlm.nih.gov/38778762/\" target=\"_blank\"\n title=\"Nucleosomal asymmetry: a novel mechanism to regulate nucleosome function\">38778762</a><br />\n [2] Lowary & Widom <em>J. Mol. Biol.</em> (1998). PMID:\n <a href=\"https://pubmed.ncbi.nlm.nih.gov/9514715/\" target=\"_blank\"\n title=\"New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning\">9514715</a><br />\n [3] Bryan et al. <em>bioRxiv</em> (2021). DOI:\n <a href=\"https://www.biorxiv.org/content/10.1101/2021.02.08.430127v1\" target=\"_blank\"\n title=\"Nucleosomal Asymmetry Shapes Histone Mark Binding and Promotes Poising at Bivalent Domains\">10.1101/2021.02.08.430127</a><br />\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Documents & Resources</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-documents\">\n <a href=\"/content/documents/tds/16-0408.pdf\" target=\"_blank\" class=\"product-documents__link\">\n <svg version=\"1.1\" id=\"Layer_1\" xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\" x=\"0px\" y=\"0px\" viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\" xml:space=\"preserve\" class=\"product-documents__icon\"\n alt=\"16-0408 Datasheet\">\n <g>\n <path class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect x=\"20\" y=\"112\" class=\"product-documents__svg-background\" width=\"146\" height=\"76\" />\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path class=\"product-documents__svg-pdf\" d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Technical Datasheet</span>\n </a>\n </div>\n </div>\n </div>\n </div>\n </div>\n</div>","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$525.00","value":525,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"Heterotypic [H3 • H3K4me3] Recombinant Nucleosome, Biotinylated","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"options":[],"related_products":[{"id":1166,"sku":"16-0410","name":"Heterotypic [H3 • H3K4me3,K14ac] Recombinant Nucleosome, Biotinylated","url":"https://www.epicypher.com/products/nucleosomes/modified-designer-nucleosomes-dnucs/heterotypic-h3-h3k4me3-k14ac-recombinant-nucleosome-biotinylated","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes","Nucleosomes/Modified Designer Nucleosomes (dNucs™)"],"summary":"\n \n \n Species: Human\n \n \n Source: E. coli & synthetic DNA\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1166/1238/Heterotypic_Nuc_white__67835.1723235454.png?c=2","alt":"Heterotypic [H3 • H3K4me3,K14ac] Recombinant Nucleosome, Biotinylated"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1166/1238/Heterotypic_Nuc_white__67835.1723235454.png?c=2","alt":"Heterotypic [H3 • H3K4me3,K14ac] Recombinant Nucleosome, Biotinylated"}],"date_added":"9th Aug 2024","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1331,"name":"Pack Size","value":"25 µg"}],"num_reviews":null,"weight":{"formatted":"0.00 LBS","value":0},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=1166","price":{"without_tax":{"currency":"USD","formatted":"$525.00","value":525},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1166"},{"id":1167,"sku":"16-0411","name":"Heterotypic [H3K4me3 • H3K14ac] Recombinant Nucleosome, Biotinylated","url":"https://www.epicypher.com/products/nucleosomes/modified-designer-nucleosomes-dnucs/heterotypic-h3k4me3-h3k14ac-recombinant-nucleosome-biotinylated","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes","Nucleosomes/Modified Designer Nucleosomes (dNucs™)"],"summary":"\n \n \n Species: Human\n \n \n Source: E. coli & synthetic DNA\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1167/1239/Heterotypic_Nuc_white__99511.1723235561.png?c=2","alt":"Heterotypic [H3K4me3 • H3K14ac] Recombinant Nucleosome, Biotinylated"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1167/1239/Heterotypic_Nuc_white__99511.1723235561.png?c=2","alt":"Heterotypic [H3K4me3 • H3K14ac] Recombinant Nucleosome, Biotinylated"}],"date_added":"9th Aug 2024","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1332,"name":"Pack Size","value":"25 µg"}],"num_reviews":null,"weight":{"formatted":"0.00 LBS","value":0},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=1167","price":{"without_tax":{"currency":"USD","formatted":"$525.00","value":525},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1167"},{"id":1168,"sku":"16-0415","name":"Heterotypic [H3K4me3 • H3K9,14,18ac] Recombinant Nucleosome, Biotinylated","url":"https://www.epicypher.com/products/nucleosomes/modified-designer-nucleosomes-dnucs/heterotypic-h3k4me3-h3k9-14-18ac-recombinant-nucleosome-biotinylated","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes","Nucleosomes/Modified Designer Nucleosomes (dNucs™)"],"summary":"\n \n \n Species: Human\n \n \n Source: E. coli & synthetic DNA\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1168/1240/Heterotypic_Nuc_white__85489.1723235678.png?c=2","alt":"Heterotypic [H3K4me3 • H3K9,14,18ac] Recombinant Nucleosome, Biotinylated"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1168/1240/Heterotypic_Nuc_white__85489.1723235678.png?c=2","alt":"Heterotypic [H3K4me3 • H3K9,14,18ac] Recombinant Nucleosome, Biotinylated"}],"date_added":"9th Aug 2024","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1333,"name":"Pack Size","value":"25 µg"}],"num_reviews":null,"weight":{"formatted":"0.00 LBS","value":0},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=1168","price":{"without_tax":{"currency":"USD","formatted":"$525.00","value":525},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1168"},{"id":1165,"sku":"16-0409","name":"Heterotypic [H3 • H3K14ac] Recombinant Nucleosome, Biotinylated","url":"https://www.epicypher.com/products/nucleosomes/modified-designer-nucleosomes-dnucs/heterotypic-h3-h3k14ac-recombinant-nucleosome-biotinylated","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes","Nucleosomes/Modified Designer Nucleosomes (dNucs™)"],"summary":"\n \n \n Species: Human\n \n \n Source: E. coli & synthetic DNA\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1165/1237/Heterotypic_Nuc_white__35995.1723235305.png?c=2","alt":"Heterotypic [H3 • H3K14ac] Recombinant Nucleosome, Biotinylated"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1165/1237/Heterotypic_Nuc_white__35995.1723235305.png?c=2","alt":"Heterotypic [H3 • H3K14ac] Recombinant Nucleosome, Biotinylated"}],"date_added":"9th Aug 2024","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1330,"name":"Pack Size","value":"25 µg"}],"num_reviews":null,"weight":{"formatted":"0.00 LBS","value":0},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=1165","price":{"without_tax":{"currency":"USD","formatted":"$525.00","value":525},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1165"},{"id":296,"sku":null,"name":"Nucleosome, Recombinant Human, H3K4me3 dNuc, Biotinylated","url":"https://www.epicypher.com/products/nucleosomes/nucleosome-recombinant-human-h3k4me3-dnuc-biotinylated","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes","Nucleosomes/Modified Designer Nucleosomes (dNucs™)","Nucleosomes/Modified Designer Nucleosomes (dNucs™)/dNuc - Smaller Pack Size Available"],"summary":"\n \n \n Species: Human\n \n \n Source: E. coli & synthetic DNA\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/296/283/dNucs__33084.1569012494.png?c=2","alt":"Nucleosome, Recombinant Human, H3K4me3 dNuc, Biotinylated"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/296/275/3_Nucleosome__08589.1569012494.jpg?c=2","alt":"Nucleosome, Recombinant Human, H3K4me3 dNuc, Biotinylated"},{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/296/283/dNucs__33084.1569012494.png?c=2","alt":"Nucleosome, Recombinant Human, H3K4me3 dNuc, Biotinylated"}],"date_added":"11th May 2016","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":null,"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$575.00","value":575},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$295.00","value":295},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$575.00","value":575},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=296"}],"shipping_messages":[],"rating":0,"meta_keywords":"H3 • H3K4me3, H3/H3K4me3, H3K4me3, heterotypic nucleosome, designer nucleosome, dNuc, nucleosome, heterotypic nucleosomes, asymmetric nucleosome, asymmetric nucleosomes","show_quantity_input":1,"title":"Heterotypic [H3 • H3K4me3] Recombinant Nucleosome, Biotinylated","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1164/1236/Heterotypic_Nuc_white__46729.1723235178.png?c=2","alt":"Heterotypic [H3 • H3K4me3] Recombinant Nucleosome, Biotinylated"}]} Pack Size: 25 µg
- Species: Human
- Source: E. coli & synthetic DNA
- Tag: Biotinylated
- Molecular Weight: 199,689 Da
Description
Heterotypic nucleosomes, also referred to as “asymmetric nucleosomes,” contain sister histones with distinct histone variants and/or post-translational modifications (PTMs). In homotypic nucleosomes, or “symmetric nucleosomes,” each pair of sister histones bears the same PTM, set of PTMs, or histone variant. Histone-modifying enzymes, chromatin remodelers, and histone chaperones differentially modify sister histones or exchange unique histone variants to form heterotypic nucleosomes. Heterotypic nucleosomes have been found at promoters of developmental genes in undifferentiated embryonic stem cells and transcription start sites (TSSs) of approximately half of the genes in budding yeast [1]. Heterotypic nucleosomes represent an additional layer of the histone code, acting as substrates for multivalent reader proteins, participating in PTM crosstalk mechanisms, and influencing reader protein binding affinity through varying local target concentration. Recombinant heterotypic nucleosomes are useful for studying chromatin dynamics and transcriptional regulation.
Heterotypic [H3 • H3K4me3] Recombinant Nucleosome, Biotinylated is a fully defined semi-synthetic nucleosome containing one unmodified histone H3 and one histone H3 with trimethylated lysine at position four. [H3 • H3K4me3] nucleosome consists of 147 base pairs of 601 sequence DNA [2] wrapped around an octamer of core histone proteins (two each of H2A, H2B, H3.2, and H4) to form a nucleosome, the basic repeating unit of chromatin. The DNA contains a 5’ biotin-TEG group, and histone H3.2 has a Cys to Ala substitution at position 110. Heterotypic [H3 • H3K4me3] nucleosomes fail to recruit known H3K4me3 binding proteins, including members of the TFIID, SETD1, and SIN3A/B complexes; chromatin remodelers NURF, CHD1, and CHD8; and PHD finger protein PHF2, suggesting that heterotypic nucleosomes may regulate transcription by affecting the recruitment of various chromatin regulatory complexes [3].
Validation Data
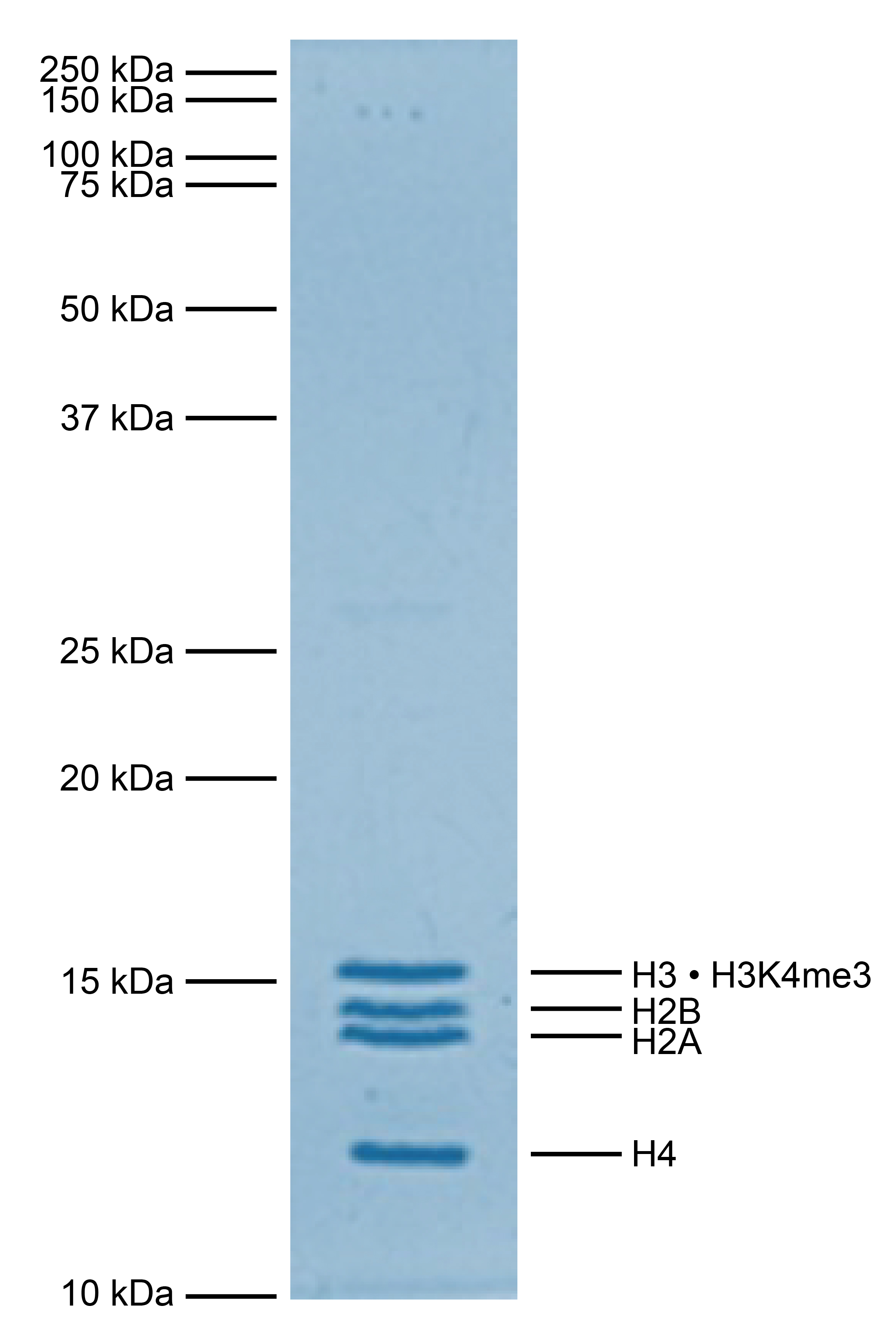
Figure 1: Protein gel data
Coomassie stained SDS-PAGE gel of proteins in heterotypic [H3 • H3K4me3] nucleosome (1 µg)
demonstrates the purity of histones in the preparation. Sizes of molecular weight markers and
positions of the core histones (H2A, H2B, H3 • H3K4me3, and H4) are indicated.
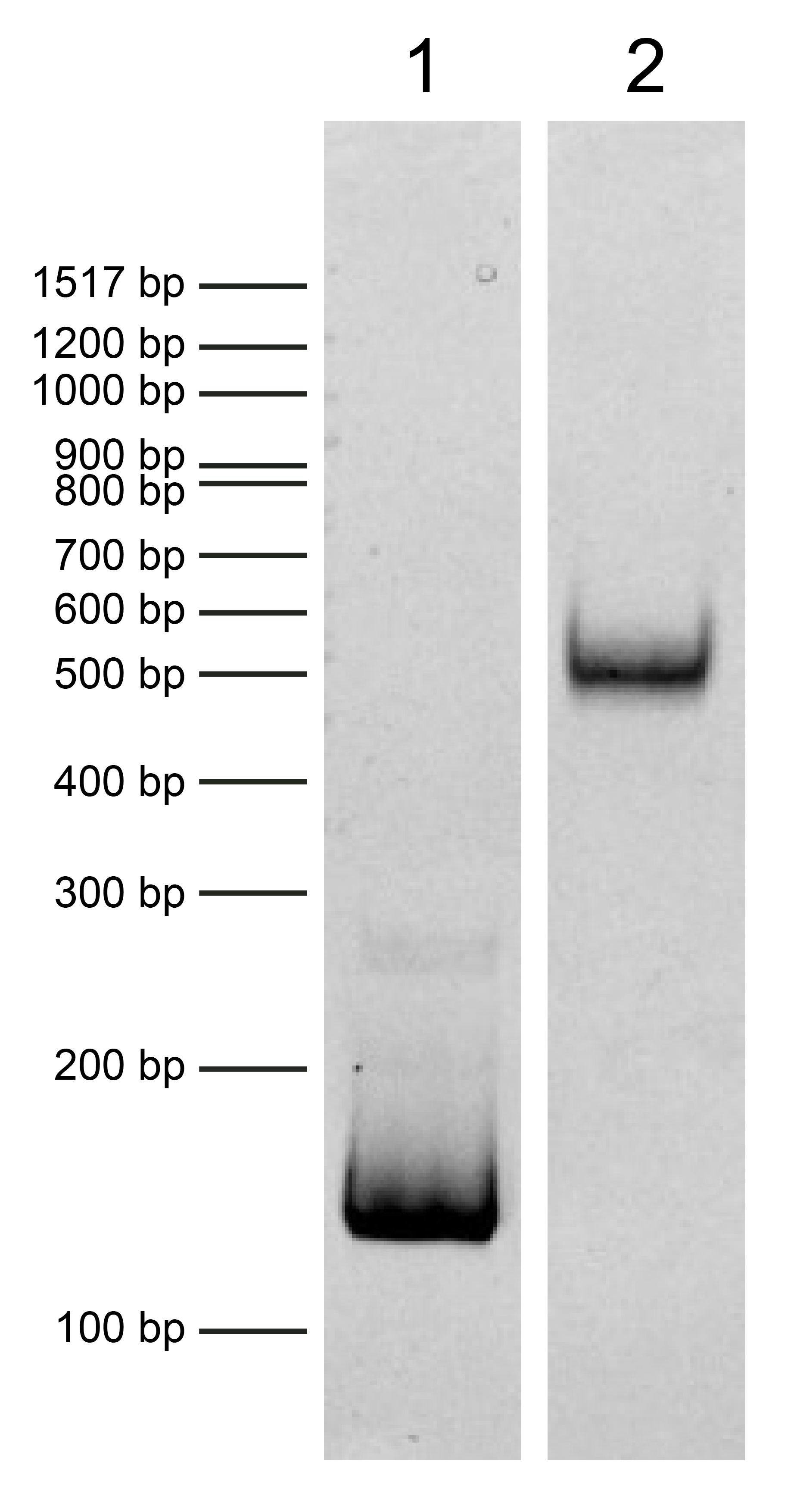
Figure 2: DNA gel data
Heterotypic [H3 • H3K4me3] nucleosome resolved via native PAGE and stained with ethidium bromide to
visualize DNA. Both lanes are from the same gel. Lane 1: Free DNA (EpiCypher 18-0005; 75 ng). Free
DNA is over 95% pure by densitometry. Lane 2: Intact heterotypic [H3 • H3K4me3] nucleosomes
(400
ng).
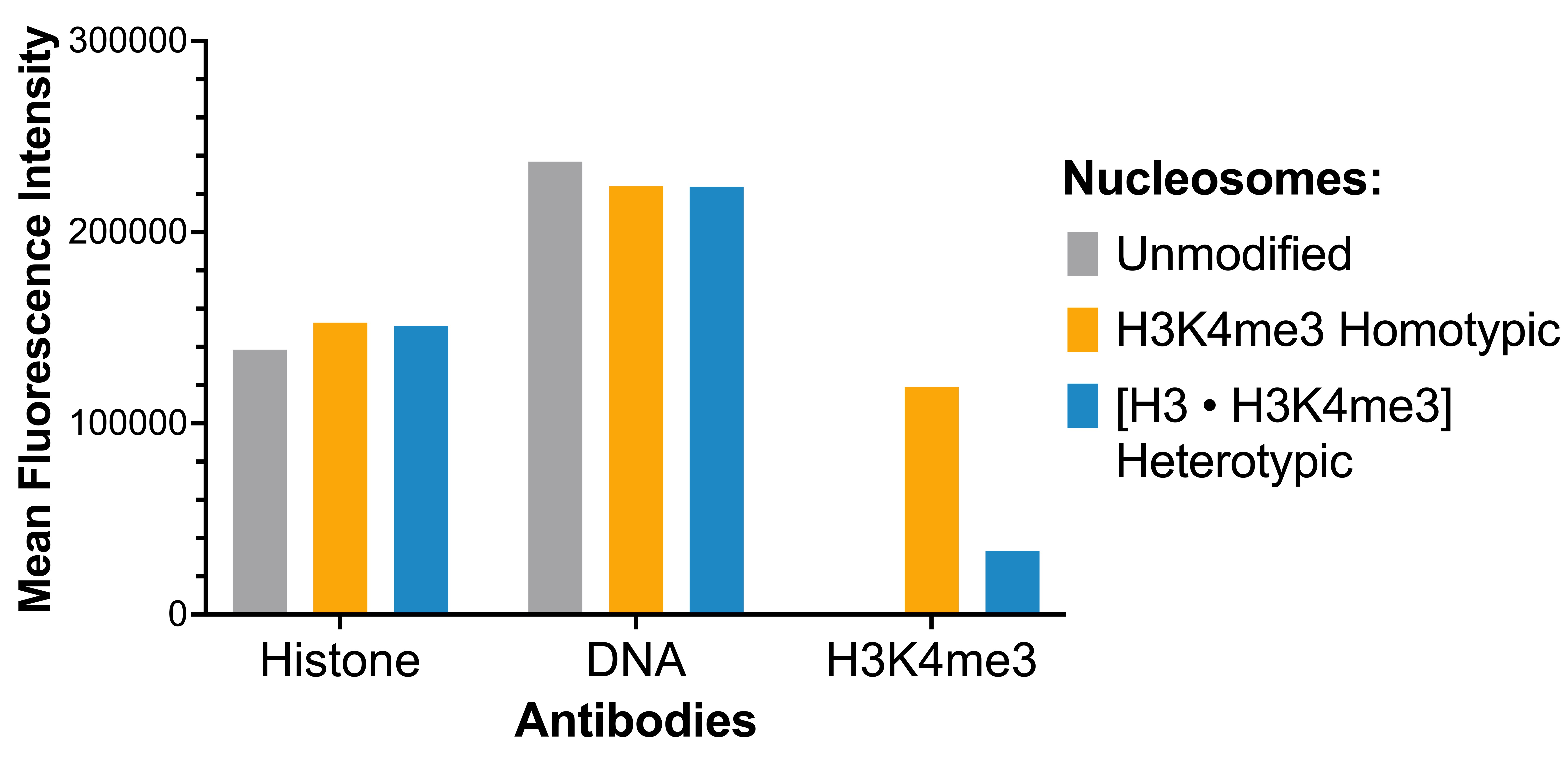
Figure 3: Luminex multiplexed specificity profiling
Heterotypic [H3 • H3K4me3] nucleosome was assessed using a Luminex® based approach. The panel
comprises biotinylated designer nucleosomes individually coupled to color-coded Luminex MagPlex®
beads. Histone antibody (EMD Millipore MAB3422), DNA antibody (EMD Millipore MAB030), and H3K4me3
antibody (EpiCypher 13-0060) were added (x-axis), and a second layer of anti-IgG*Phycoerythrin
(PE; BioLegend 406421 for anti-rabbit IgG or BioLegend 405307 for anti-mouse IgG) was used for
detection. Data were generated using a Luminex FlexMAP 3D®. Histone and DNA antibodies bind
nucleosomes in the panel equivalently, as expected. H3K4me3 antibody binds heterotypic [H3 •
H3K4me3] nucleosomes at reduced strength in comparison to homotypic H3K4me3 nucleosomes (EpiCypher
16-0316).
Technical Information
Application Notes
Heterotypic [H3 • H3K4me3] nucleosome is highly purified and suitable for a variety of applications, including use as a substrate in enzyme assays, high-throughput screening and inhibitor testing, chromatin binding studies, protein-protein interaction assays, structural studies, and in effector protein binding experiments. Trace amounts of TEV protease may be present (less than 5% of the preparation by densitometry, Figure 1). For a corresponding homotypic control, we recommend EpiCypher 16-0316.
Gene & Protein Information
H2B - O60814 (alt. names: H2B K, HIRA-interacting protein 1)
H3.2 - Q71DI3
H4 - P62805
References
[1] Valsakumar & Voigt. Biochem. Soc. Trans. (2024). PMID: 38778762
[2] Lowary & Widom J. Mol. Biol. (1998). PMID: 9514715
[3] Bryan et al. bioRxiv (2021). DOI: 10.1101/2021.02.08.430127

![Heterotypic [H3K4me3 • H3K14ac] Recombinant Nucleosome, Biotinylated](https://cdn11.bigcommerce.com/s-y9o92/images/stencil/500x500/products/1167/1239/Heterotypic_Nuc_white__99511.1723235561.png?c=2)
![Heterotypic [H3K4me3 • H3K9,14,18ac] Recombinant Nucleosome, Biotinylated](https://cdn11.bigcommerce.com/s-y9o92/images/stencil/500x500/products/1168/1240/Heterotypic_Nuc_white__85489.1723235678.png?c=2)
![Heterotypic [H3 • H3K14ac] Recombinant Nucleosome, Biotinylated](https://cdn11.bigcommerce.com/s-y9o92/images/stencil/500x500/products/1165/1237/Heterotypic_Nuc_white__35995.1723235305.png?c=2)