Nucleosome spike-ins for quantitative ChIP-Seq
Chromatin immunoprecipitation (ChIP) is a powerful and widely used approach to understanding chromatin regulation. However, ChIP experiments have numerous sources of technical challenges that can be avoided by using SNAP-ChIP® Spike-In Controls:
- Ensure antibody specificity
- Normalize ChIP data
- Test antibody enrichment
- Monitor technical variation
Have Questions?
We’re here to help. Click below and a member of our team will get back to you shortly!
Request More InfoSample Normalization & Antibody Profiling for ChIP (SNAP-ChIP) spike-in controls provide a powerful new tool to generate highly reliable ChIP-seq data
What are SNAP-ChIP® Spike-in Controls?
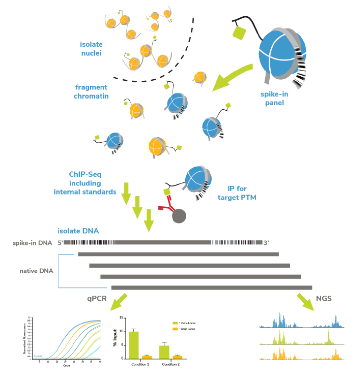
SNAP-ChIP spike-in controls are panels of recombinant nucleosomes carrying widely studied histone post-translational modifications (PTMs), each denoted by a unique DNA barcode.
SNAP-ChIP spike-in control panels are compatible with both native and cross-linked workflows. Simply spike-in the panel of interest prior to IP.
The nucleosomal DNA barcodes on the spike-ins can then be detected by qPCR or NGS and used to quantify antibody performance and normalize samples for reliable cross-experimental comparisons.
Use SNAP-ChIP Spike-Ins To Control Your ChIP Experiment
Ensure Antibody Specificity
Our findings1 and several publications have found that:
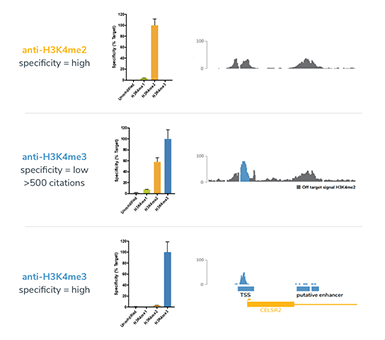
- Histone peptide arrays used for antibody validation do not predict specificity in ChIP1.
- Antibody specificity can vary significantly between production lots (see our blog).
- The use of highly cited antibodies does not guarantee quality results1,2.
- Many ChIP-grade antibodies cross-react, leading to incorrect biological interpretations (see figure)1,2.
Only by validating with SNAP-ChIP spike-in controls can scientists correctly identify and avoid cross-reactive antibodies which would confound their results.
Monitor Technical Variation
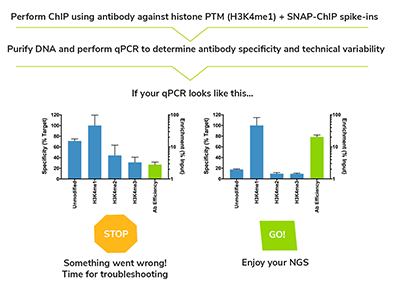
Our spike-in controls will make sure you never sequence another failed ChIP experiment again.
- Seamlessly integrate into native and cross-linked ChIP workflows.
- Serve as positive (on-target) and negative (off-target) controls.
- Monitor technical variability in antibody specificity / efficiency across replicates.
- qPCR of DNA barcodes provides a valuable STOP / GO step prior to expensive sequencing
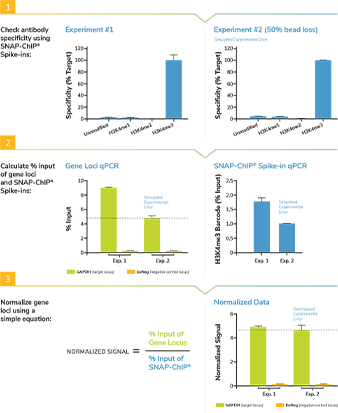
Normalize Between Experiments
SNAP-ChIP spike-in controls can also be used to normalize between experiments3,4.
Some methods use exogenous chromatin (e.g. Drosophila) for normalization. However, these approaches are problematic because they are heterogeneous, poorly defined, and inconsistent between lots.
SNAP-ChIP spike-ins provide a homogeneous defined control and are consistent from lot to lot. EpiCypher is currently working to create more robust and streamlined SNAP-ChIP workflows for ChIP normalization.
Interested in SNAP-ChIP Spike-in Controls?
SNAP-ChIP® Spike-In Control Panel
Defined controls for superior ChIP. We offer panels for a variety of histone PTMs, including lysine methylation and acylation.
SNAP-ChIP® Certified Antibodies
These antibodies have been validated using SNAP-ChIP spike-ins and have best-in-class specificity and target enrichment.
SNAP-ChIP® Validation Service
Let the experts help! With SNAP-ChIP Validation Services, EpiCypher will help find the best antibody for your ChIP-seq studies.
Featured Publications
1. Shah et al. Examining the roles of H3K4 methylation states with systematically characterized antibodies. Mol Cell. 72, 162 - 177 (2018). (PMID: 30244833)
In this study, SNAP-ChIP Spike-in technology revealed that many widely cited, “ChIP-grade” antibodies exhibit poor binding specificity and pull-down efficiency, and demonstrated that histone peptide arrays do not accurately predict antibody performance.
2. Lam et al. Cell-type-specific genomics reveals histone modification dynamics in mammalian meiosis. Nat. Commun. 10, 3821 (2019). (PMID: 31444359)
SNAP-ChIP K-MetStat spike-ins were used to normalize ChIP-seq data and uncover epigenetic changes associated with meiosis. In choosing an antibody for their study, Lam and colleagues recapitulated findings from Shah et al., in which widely cited H3K4me3 antibody produced biologically distinct findings compared to a highly specific EpiCypher SNAP-ChIP Certified H3K4me3 antibody.
3. Grzybowski et al. Native internally calibrated chromatin immuneprecipitation for quantitative studies of histone post - translational modifications. Nat. Protoc. 14,3275-3302 (2019). (PMID: 31723301)
This study includes detailed methods for the use of barcoded recombinant nucleosomes as spike-in controls for ChIP normalization, and are directly applicable to the use of SNAP-ChIP spike-ins.
4. Tay et al. Hdac3 is an epigenetic inhibitor of the cytotoxicity program in CD8 T cells. J. Exp. Med. 217, e20191453 (2020). (PMID: 32374402)
Reads from the DNA barcodes on SNAP-ChIP K-AcylStat spike-ins were used to normalize H3K27ac ChIP-seq data and elucidate H3K27ac changes after HDAC3 knockout in CD8+ T cells.
5. Shirane et al. NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nature Genetics 52, 1088-1098 (2020). (PMID: 32929285)
Here, the Lorincz lab used SNAP-ChIP Certified Antibodies and Spike-in Controls to characterize the histone lysine methylation landscape in mouse male germline cells. These studies revealed a novel role for the H3K36 methyltransferase NSD1 in driving sexually dimorphic DNA methylation patterns in the germline.