Defined Spike-in Controls and a Validated Workflow for our New CUT&RUN Kits
CUT&RUN is quickly becoming a leading assay for chromatin profiling, and it’s easy to see why. The targeted cleavage of antibody-bound chromatin allows for streamlined sample processing and greater signal : noise, meaning researchers can generate high quality data with fewer cells and sequencing reads. EpiCypher’s CUTANA™ ChIC / CUT&RUN Kit comes with nearly everything you will need for a successful CUT&RUN experiment, and each reagent has been meticulously validated and re-validated, from the pAG-MNase enzyme down to the included DNA purification columns.
Our thorough kit optimization also resulted in the development of a robust CUT&RUN protocol, validated for diverse targets, sample types, as well as low cell inputs (see below). Furthermore, the CUTANA CUT&RUN Kit comes with all the necessary experimental controls, including positive and negative control antibodies, E. coli spike-in DNA for data normalization, and a panel of DNA-barcoded recombinant modified designer nucleosomes (dNucs™) to monitor the success of your CUT&RUN experiment. Combined, these features make our CUTANA CUT&RUN Kit a standout tool for accurate and user-friendly chromatin profiling.
Learn more about CUTANA™ ChIC / CUT&RUN Assays
CUTANA™ CUT&RUN Kit provides all necessary controls for your CUT&RUN experiment
A well designed, publication-quality experiment always includes the appropriate experimental controls. Our CUTANA CUT&RUN Kit provides multiple controls and clear instructions for their use in CUT&RUN, as well as how to draw conclusions from the quality checkpoints before sequencing and at data analysis. Here we review the types of controls included in the CUTANA CUT&RUN Kit, and how these controls make EpiCypher’s kit a top choice for your next CUT&RUN experiment.
Positive and negative control antibodies: The CUT&RUN Kit contains positive (rabbit anti-H3K4me3) and negative (rabbit anti-IgG) control antibodies, which are added to control reactions at the same time as antibodies to your target of interest are added to the experimental samples. Our H3K4me3 antibody has been confirmed to exhibit high specificity and efficiency in chromatin immunoprecipitation (ChIP) by using defined nucleosome controls (i.e. using SNAP-ChIP® Spike-ins)1.
For more information, check out our H3K4me3 positive control antibody blog post.
Unlike any other antibody available on the market, every lot is also tested in CUT&RUN and shown to exhibit strong correlation with ChIP-seq data. This exceptional focus on lot testing, specificity determination using defined physiological controls, and platform specific validation go above and beyond to aid in experimental rigor and reproducibility, thus providing a best-in-class antibody for establishing CUT&RUN assays in your lab.
In your experiment, the tracks from the control antibody should produce the expected distribution and peak structure in the species tested, and should be consistent across replicates. The negative control antibody tracks reveal background signal and the positive control antibody tracks indicate the success of the workflow (see Figure 1).
dNuc spike-in controls: monitor experimental success on a defined chromatin substrate. Exclusive to the EpiCypher CUT&RUN Kit are DNA-barcoded dNuc controls, designed to work with the positive control H3K4me3 antibody. The CUTANA H3K4 MetStat Spike-in Controls (H3K4me0, H3K4me1, H3K4me2, and H3K4me3 dNucs) are wrapped with DNA that contains a biotin tag, allowing them to be coupled to streptavidin magnetic beads (SA Beads, Figure 2A). These dNuc-beads are spiked in with ConA-bead immobilized nuclei, just prior to the addition of the H3K4me3 control antibody in the experimental workflow. Targeted cleavage with pAG-MNase releases linker DNA from antibody-bound H3K4me3 dNuc (but not the other off-target dNucs) into solution, where it is collected for library preparation and sequencing analysis.
Each nucleosome has a unique 22 bp barcode sequence for identification post-sequencing. The relative sequencing read counts from each dNuc control allows the user to quantitatively assess antibody specificity, thereby gauging experimental success and guiding troubleshooting efforts for problematic experiments. Expected results from the CUTANA H3K4 MetStat Panel are shown in Figure 2B, using EpiCypher’s entire panel of H3K4me-state antibodies as well as the IgG negative control antibody. For comparison, we also tested a widely cited H3K4me3 antibody previously found to exhibit substantial H3K4me2 cross-reactivity in ChIP-seq1 (see right column, Figure 2B). This antibody cross-reacted with H3K4me2 in CUT&RUN, illustrating the power of our dNuc controls to flag improperly characterized reagents for this novel chromatin profiling assay.
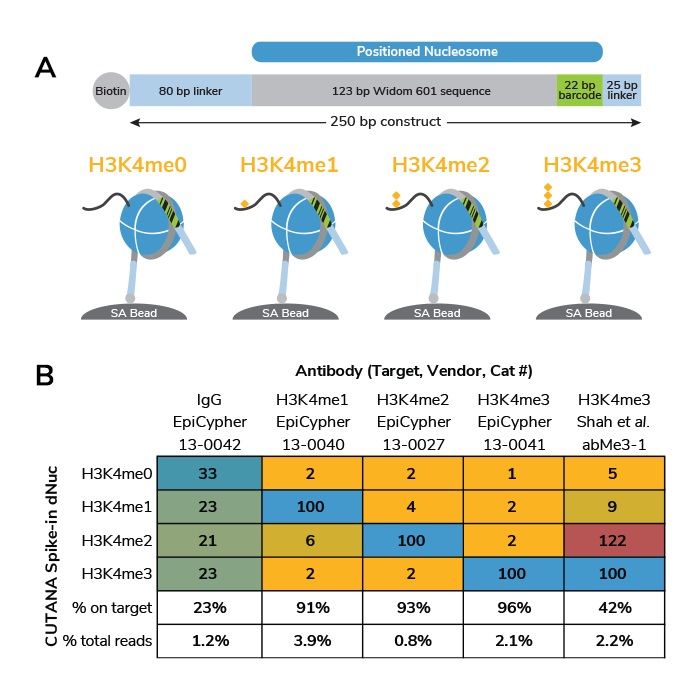
But how do nucleosome spike-ins work as a control? The use of the CUTANA dNuc spike-in panel together with the positive and negative control antibodies allows researchers to quantitatively evaluate their CUT&RUN technique. We recommend both new and experienced users perform control reactions with these dNucs and provided control antibodies in every experiment. If off-target PTM cross-reactivity exceeds ~5% of the on-target reads (in a native workflow), then the user should follow suggested troubleshooting steps as outlined in our manual.
E. coli DNA spike-in: Control for library prep and normalize sequencing data. Another important control included in the CUTANA CUT&RUN Kit is E. coli spike-in DNA. This sheared DNA is added to samples after pAG-MNase cleavage, and thus can be used to monitor DNA loss and/or amplification during library prep. In addition, reads from E. coli DNA spike-in can easily be used to normalize sequencing data and perform pairwise comparisons, as done for ChIP-seq2 and in recent CUT&RUN studies3 using EpiCypher’s pAG-MNase.
Compatible with diverse targets and inputs
When you order the CUTANA CUT&RUN Kit, you’re getting much more than a chromatin profiling kit- you are also gaining access to all of EpiCypher’s experience in developing and optimizing this remarkable assay. Our goal in developing this kit was to make it user-friendly, reliable, and compatible with diverse targets, sample inputs, and low cell numbers. We have met these metrics with our CUTANA CUT&RUN Kit, as shown in a series of illustrative experiments below.
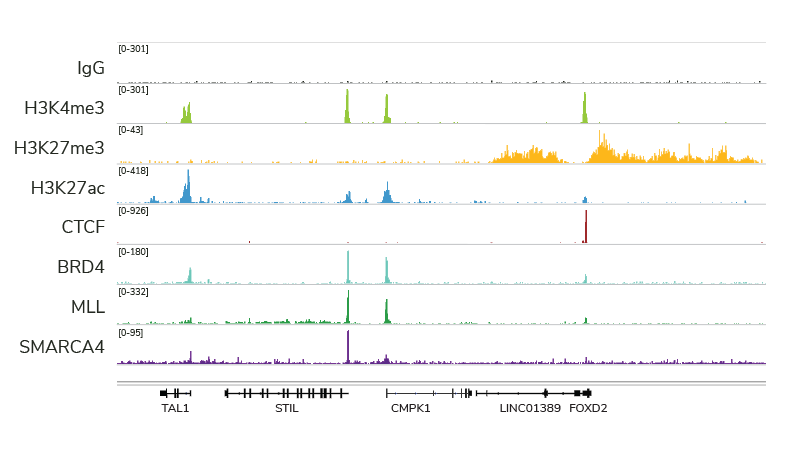
Profiling diverse chromatin-associated protein targets and histone post-translational modifications (PTMs). As shown in Figure 3, EpiCypher’s CUTANA pAG-MNase enzyme and kit have been validated for a broad array of chromatin interacting proteins, including transcription factors (e.g. CTCF), epigenetic regulators (e.g. BRD4), chromatin writer enzymes (e.g. MLL), and even transiently-interacting chromatin remodeling proteins (e.g. SMARCA4, historically difficult to ChIP). Importantly, each of these datasets was generated using the standard native CUT&RUN approach with a sequencing depth of 3-8 million reads, a fraction of the typical depth required for ChIP-seq (>30 million reads).
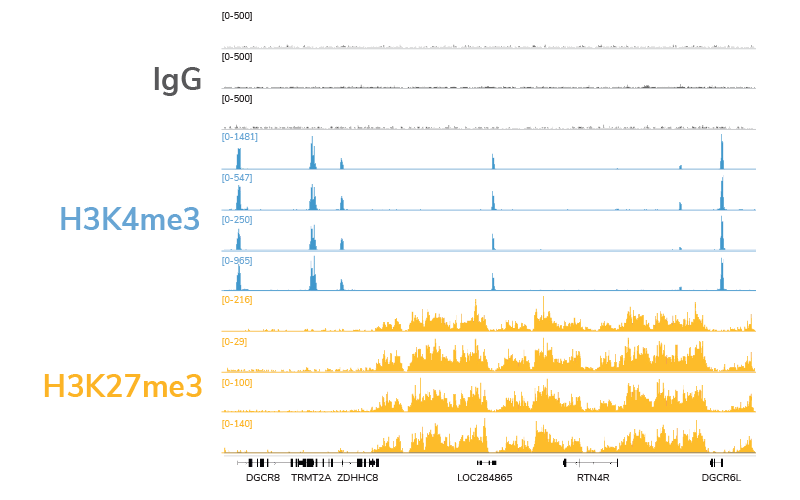
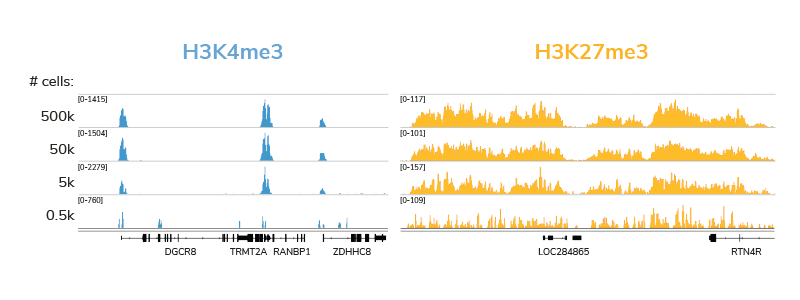
Compatibility with low cell inputs. A major advantage of CUT&RUN over ChIP-seq is compatibility with low cell numbers, making it an ideal approach for experiments where samples are limited (e.g. clinical applications). Our extensively validated CUTANA CUT&RUN protocol and kit are optimized for 500,000 cells, but generate comparable data down to 5,000 cells without any adaptations to the workflow. Figure 4 shows representative genome browser tracks from CUT&RUN cell titrations using antibodies against two distinct classes of histone PTMs: H3K4me3, a low-abundance target that occurs in sharp peaks at active transcription start sites, and H3K27me3, a high-abundance target that occurs in broad peaks in facultative heterochromatin. All data from these experiments display high signal : noise and low background, and are compatible with 3-8 million reads / sample.
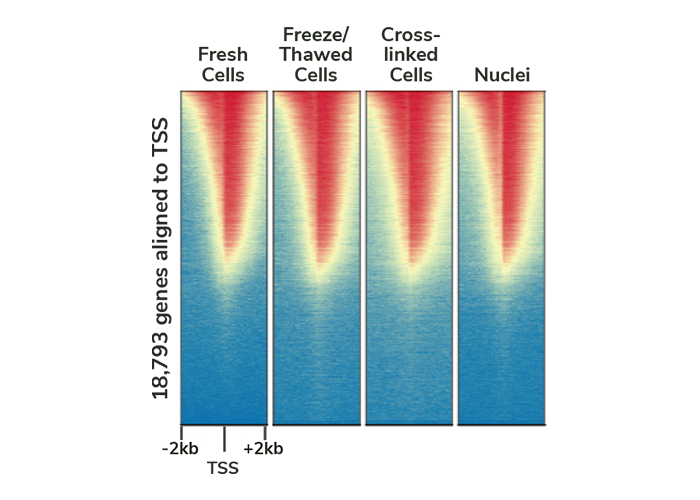
Can be adapted for fixed / frozen samples. The original CUT&RUN assay was designed specifically for native (i.e. unfixed) chromatin4-6. While native CUT&RUN works well for most targets, including many transiently-interacting transcription factors and chromatin remodelers, some targets (e.g. labile lysine acetylation marks) may be stabilized and preserved by cross-linking. In addition, labs often work with archived, frozen samples, or isolated nuclei, instead of fresh cells from culture.
To address these challenges and make the CUTANA CUT&RUN Kit applicable to all types of sample inputs, we developed protocol adaptations for these common chromatin preparation methods. Our objective was to deliver protocols that generate data of equal quality to that from fresh cells. We optimized these adaptations using H3K4me3 CUT&RUN and K562 cells, and results are shown in Figure 5. Our final methods meet this metric and provide consistent enrichment of H3K4me3 across all sample preparation protocols. However, because cross-linking reduces yield and can introduce artifacts, we recommend always starting with native CUT&RUN first when possible, as this works well for the majority of targets.
Order CUTANA™ CUT&RUN Kit and get started with CUT&RUN today
EpiCypher’s CUTANA CUT&RUN Kit represents a breakthrough advance in high quality chromatin profiling, with every detail thoroughly tested for user success. Order your CUT&RUN Kit now or, if you have any questions, feel free to reach out to us using the contact form below.
References
- Shah RN et al. Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies. Mol Cell 72, 162-77 e7 (2018). PMID:30244833
- Orlando DA et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 9, 1163-70 (2014). PMID:25437568
- Tay RE et al. Hdac3 is an epigenetic inhibitor of the cytotoxicity program in CD8 T cells. J Exp Med 217, (2020). PMID:32374402
- Meers MP et al. Improved CUT&RUN chromatin profiling tools. Elife 8, (2019). PMID:31232687
- Skene PJ et al. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 13, 1006-19 (2018). PMID:29651053
- Skene PJ et al. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, (2017). PMID:28079019
