Will my sample work in CUT&RUN? Four factors to consider

CUT&RUN assays use intact cells or nuclei to map the genome-wide enrichment of histone post-translational modifications (PTMs) and chromatin-associated proteins, such as transcription factors. At EpiCypher, we are often asked, “Will my cell samples work for CUT&RUN?” Although it may be tempting to assume that all cells will perform similarly, this is not the case.
Here we will discuss the main criteria EpiCypher uses when assessing cells for CUT&RUN experiments. In general, remember that bad sample quality = bad CUT&RUN data.
Ready to start CUT&RUN? See the CUTANA™ CUT&RUN Kit
1. Cell Number
EpiCypher recommends using 500,000 cells per reaction for robust CUT&RUN profiles. We count cells at two steps in the workflow: once at the initial cell harvest and again before immobilizing cells to magnetic beads. The second step ensures that you haven’t lost significant sample or incurred cell lysis during washes with CUT&RUN buffers. See our Tech Support Center for specific guidance.
CUTANA™ CUT&RUN is successful down to 5,000 cells for select targets. Keep in mind that using lower cell numbers may reduce CUT&RUN yields, necessitating adjustments in library prep and sequencing. In these cases, see our CUT&RUN Library Prep Kit manual for helpful suggestions.
2. Viability
EpiCypher determines cell viability, or the percentage of cells that are “live,” at the initial cell harvest. The viability of starting cells is critical for successful CUT&RUN. Low viability can contribute to increased assay background, low yields, and problems with cell/bead clumping during the protocol.
Keep in mind that optimal viability is highly dependent on the sample type. For instance, we aim for our K562 cells cultures to have >90% viability when harvesting for CUT&RUN. However, for some sample types or experimental conditions, the optimal viability may be lower.
Tips on Viability and cell counting methods
EpiCypher suggests using a simple Trypan Blue staining method to count cells and determine starting cell viability. Because Trypan Blue dye is toxic to cells, be sure to count cells immediately after adding the dye.
Some cells show increased sensitivity to Trypan Blue. If your cells show poor viability by Trypan Blue staining, yet have expected morphology, minimal lysis, and normal growth in culture, these cells are probably fine for CUT&RUN. We suggest using a more dilute Trypan Blue dye or trying other cell counting methods to confirm cell viability, such as Propidium Iodide.
Guidance on sample prep for CUT&RUN
3. Cell morphology and integrity
CUT&RUN requires intact cells with normal morphology. Morphology refers to cell shape and relates to the tissue source and/or how cells grow in culture. Suspension cultures, such as K562 cells, which commonly derive from blood cells and/or cancers, have round and symmetric morphology. Adherent cells can be derived from epithelial, endothelial, neuronal, or fibroblast tissues, and thus display diverse morphology and expansion characteristics. For instance, epithelial cells tend to have a uniform cell shape and grow in patches on cell culture plates, while fibroblasts show asymmetric, elongated morphology, and can be used to study cell migration.
Using lysed or poor-quality cells will lead to equally poor data. To ensure robust sample prep, EpiCypher checks morphology and cell membrane integrity at both initial cell collection and prior to bead binding. The second check is important, as some sample types (e.g. FACS-isolated cells or those from tissues) can be more sensitive to lysis in CUT&RUN Wash Buffers. In these cases, we recommend isolating nuclei for CUT&RUN instead ( Figure 1).
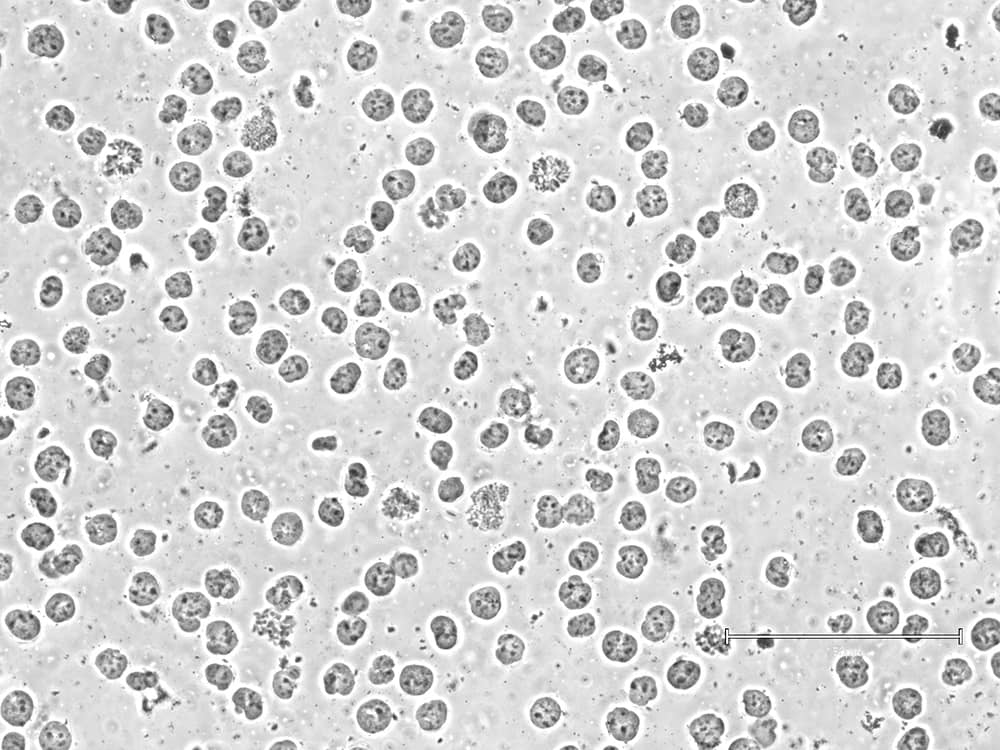
Isolate nuclei using our Nuclei Extraction Buffer!
4. Cell source considerations: cell lines, primary cells, tissues, and stem cells
Morphology, viability, and cell numbers are directly influenced by the source of your cells. CUT&RUN can be performed using cells from tissues, cell lines, cultured primary cells, or cells purified by other means (e.g. FACS). Here we discuss the advantages and challenges associated with these samples.
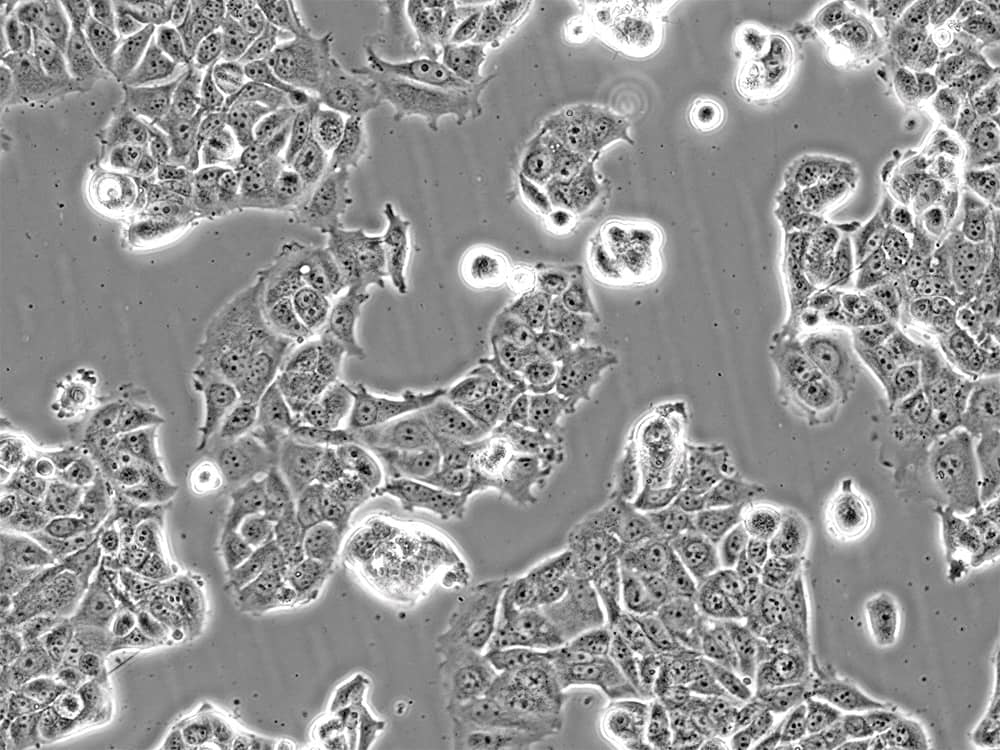
Immortalized cell lines
Examples: HEK293, HeLa, K562, NIH/3T3, MCF-7 (Figure 2), H1299 (Figure 3), A549, and THP-1 cell lines.
Advantages: In general, cell lines are the easiest inputs to optimize for CUT&RUN. Cell lines provide:
- Large quantities of cells with high viability – key for CUT&RUN success.
- Robust and highly reproducible CUT&RUN profiling, due to homogeneity of cell lines.
- Robust systems for drug screening, gene overexpression/repression, and more - without destroying sample quality.
Disadvantages: The use of cell lines comes with many caveats, outlined below. However, depending on your experimental goals, cell lines may be the best option.
- In general, cell lines are not representative of in vivo conditions. They are often derived from cancers or other diseased cell populations, and their function may be different compared to primary cells.
- Cell lines are prone to genomic alterations, including chromosomal abnormalities, duplications, and/or genetic drift.
- Contamination with other cell lines is common. Misidentification can hamper assay reproducibility and lead to incorrect biological conclusions.
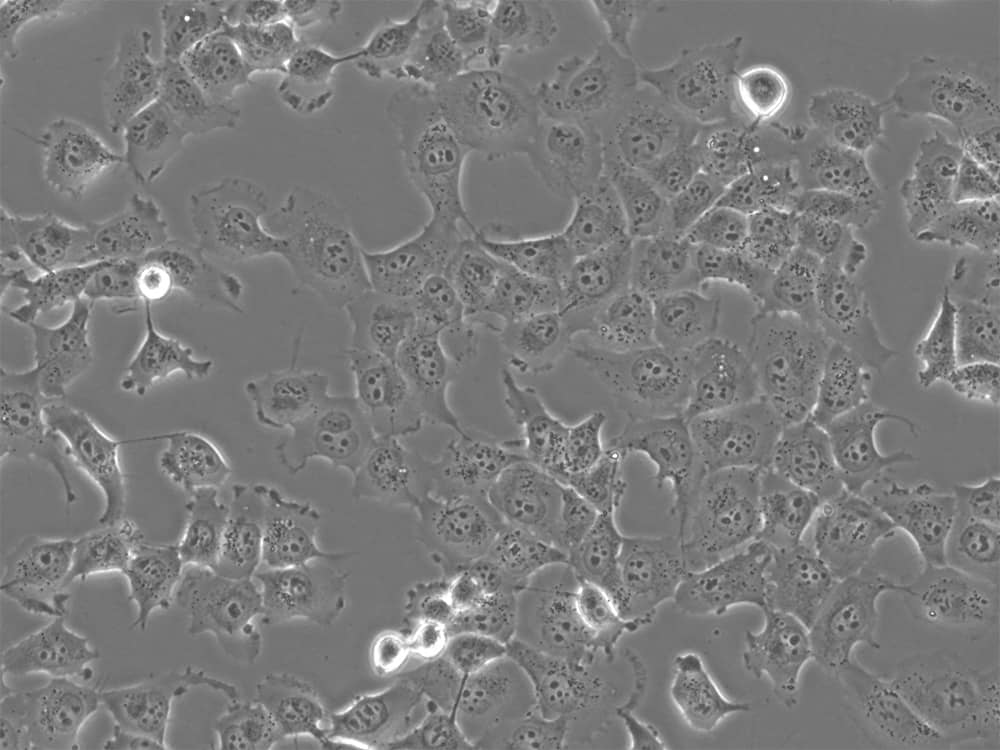
Primary cells
Examples: Samples derived from living tissues, either solid (e.g. intestine, lung, liver, skin) or fluid/liquid (e.g. blood), with or without cell culture.
Advantages: Primary cells are frequently used in CUT&RUN, as they enable scientists to:
- Examine chromatin function and biological mechanisms in a native, heterogeneous cellular environment.
- Study the function of rare cell states isolated by FACS or other techniques.
- Determine cellular responses to stimuli or drug treatments in vivo.
- Characterize patient samples (e.g. tumor vs. healthy cells).
Disadvantages: The drawbacks to primary cells rely on the type of tissue and cell type you want to study. When studying primary cells, it can be challenging to:
- Isolate adequate cell numbers, either due to cell sensitivity during isolation (e.g. FACS) or low abundance in tissues.
- Obtain cells with high viability – primary cells often require careful handling to prevent lysis.
- Expand in culture. Their ability to expand in culture is usually limited to a few passages, requiring careful planning and optimization of experimental timelines.
- Generate consistent results, especially from bulk tissues containing many distinct cell types.
Stem cells
Examples: Induced pluripotent stem cells (iPSCs), mesenchymal stem cells, and embryonic stem cells.
Advantages: The ability to grow and culture pluripotent and adult stem cell populations provides a wealth of opportunities to:
- Study epigenetic changes during cell development, differentiation, and disease progression.
- Generate rare cell types for functional analysis and therapeutic development.
- Develop patient-specific cell populations for personalized medicine applications.
- Study cell growth and differentiation using two-dimensional and/or organoid cultures.
Disadvantages: Despite their promise in cell and gene therapy research and developmental biology, working with stem cells can be challenging in many respects:
- Stem cell culture requires high-level expertise, and even then, expanding some stem cell populations is difficult.
- Culturing stem cells requires multiple quality control measures and precise monitoring, translating into expensive, time-consuming efforts.
- Differentiation protocols can vary between researchers, introducing unexpected biases and variations.
Design your experiment - visit our Tech Support Center!
Still have questions? Our Technical Support Center includes multiple protocol modifications for different cell types, including adherent cell types, immune cells, tissues, and more. These modifications have been validated with EpiCypher’s CUTANA CUT&RUN Kit and Library Prep Kit, and are also compatible with our CUT&RUN antibodies.
Our technical support team is also here to help. Please reach out and we will get back to you soon!
