Webinar recap: Nucleosomes define the histone code

At the center of chromatin regulatory mechanisms is the nucleosome – a histone octamer wound by DNA. The tails of histone proteins are decorated with chemically diverse post-translational modifications (PTMs), such as methylation and acetylation, which recruit and/or interact with various chromatin regulatory proteins to control genome structure and function. Histone PTMs are often considered a molecular language or “histone code,” due to their combinatorial binding mechanisms, and have been the subject of decades of biomedical research and drug development1. However, recent work has shown that other nucleosome features are also critical for understanding chromatin interactions and gene expression pathways. In this webinar, EpiCypher’s Chief Scientific Officer Dr. Michael-Christopher Keogh discussed why studying chromatin regulation in the nucleosomal context is essential for chromatin biology. Read on for a recap!
Watch the webinar on-demand now!
Histone peptides don’t tell the whole story
Historically, the histone code hypothesis centered on the interactions of PTMs located on N-terminal histone tails, which were thought to extend outward from the nucleosome core (as in Figure 1). Histone PTM interactions were examined using modified histone peptides, which are usually small, N-terminal segments of individual histone proteins. Although recombinant nucleosome assembly methods were available, they were expensive, technically difficult, and generated nucleosomes of variable quality. In contrast, highly pure modified histone peptides could be generated at-scale using automated protein synthesis procedures, providing defined substrates for binding or enzymatic assays. Histone peptide microarrays were the standard approach for defining histone PTM binding specificity, due to their low production cost, ease of use, and widespread availability1.
However, histone peptides alone fail to reveal the whole story.
To exert their regulatory roles, many chromatin-associated proteins engage histone PTMs and other nucleosome components, such as DNA or DNA modifications (e.g. 5mC) and the nucleosome surface (e.g. H2A/H2B acidic patch) – a concept known as multivalency. Recent NMR and structural analyses indicate that the positively charged histone tails also have high affinity for nucleosomal DNA, such that tails are collapsed onto the nucleosome core. These data contrast with classic depictions of protruding histone tails, and support the role of PTMs in complex binding mechanisms1.
As the field advances, the role of multivalency in regulating chromatin structure, interactions, and gene expression highlights the need for biologically-relevant substrates. By definition, histone peptides do not contain all the molecular pieces required to model physiological chromatin. As discussed below, peptides can result in incorrect or incomplete conclusions in studies of chromatin regulation or, as EpiCypher has found, inaccuracies in PTM antibody characterization (see this blog and chromatinantibodies.com).
To meet these needs, EpiCypher pioneered the commercial development of fully-defined, semi-synthetic/recombinant nucleosomes. These reagents contain known PTMs or mutations and have been adapted for many applications, including high-throughput EpiDyne™chromatin remodeling platforms, comprehensive dCypher™ binding screens and even as physiologically-relevant SNAP Spike-in Controls for chromatin mapping assays.
Histone PTM interplay drives chromatin binding
In his webinar, Dr. Keogh provided several examples of how nucleosome substrates are enabling key discoveries that would be obscured or impossible using histone peptides.
Before diving in, it’s important to note that in vivo, histone PTMs almost never occur in isolation. The histone octamer contains two of each core histone, in the form of two H2A/H2B dimers and one H3/H4 tetramer. Each of these histones can carry different PTMs and contribute to unique chromatin interactions. For instance, if looking at a nucleosome carrying H3K4me3 + H3K9ac, the two modifications could be on the same H3 tail (in cis) or be localized on separate H3 tails (in trans; Figure 1).
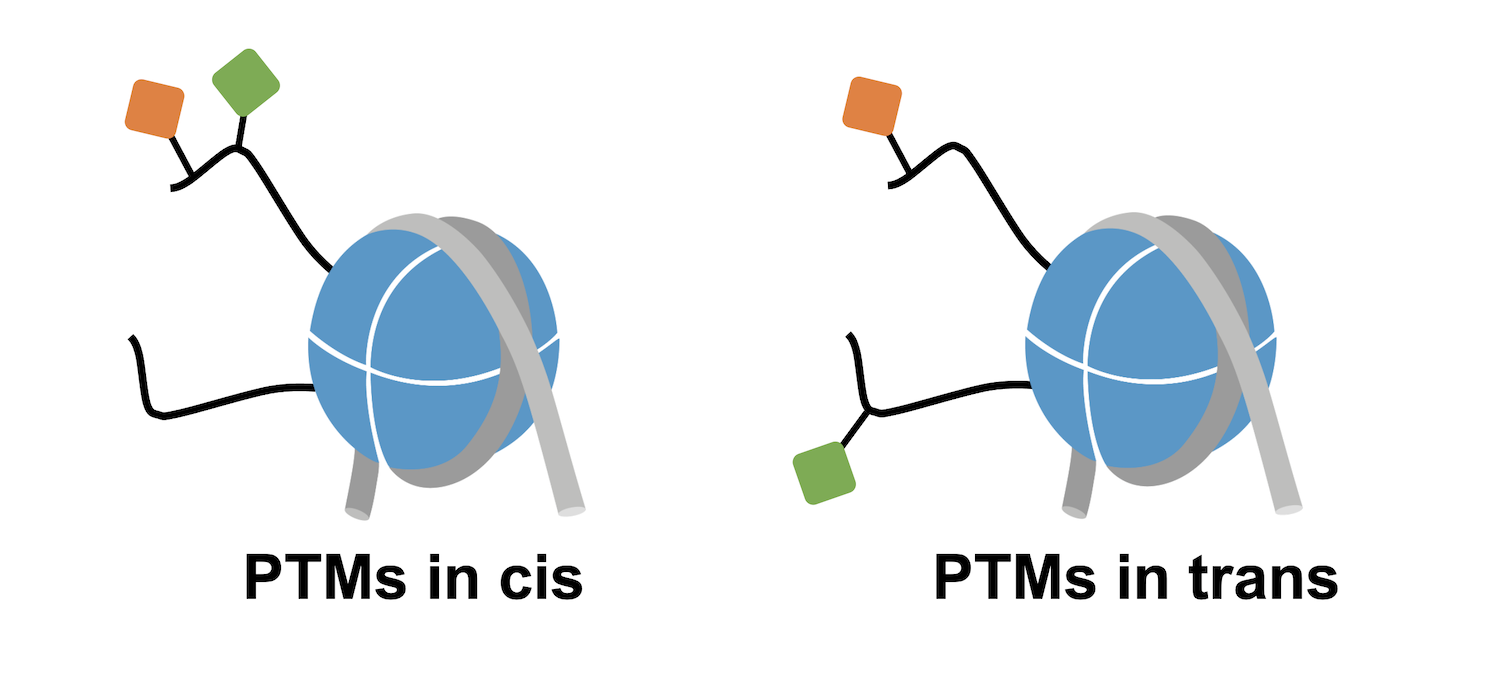
Histone PTMs also have additional functions that can impact downstream epigenetic mechanisms. In particular, histone lysine acetylation neutralizes the positive charge of histone tails, reducing the affinity of tails for negatively charged DNA and/or altering interactions with the surface of the nucleosome. Thus, acetylation can mediate changes in histone tail accessibility that effectively regulate chromatin interactions1.
These scenarios raise multiple questions about histone PTM interplay, which can only be addressed using nucleosome substrates. Below are two scientific stories discussed by Dr. Keogh that highlight the importance of nucleosome context.
BPTF
BPTF is a chromatin reader protein and subunit of the nucleosome remodeling factor (NURF) complex. BPTF contains tandem reader domains for histone lysine acetylation and methylation, consisting of a PHD finger and bromodomain (BPTF PHD-BD), providing an excellent opportunity to dissect combinatorial PTM interactions. Indeed, BPTF has been the subject of many studies using histone peptides, which suggested strong engagement with H3K4me3 and a variety of H3Kac and H4Kac marks. However, these studies ignored the potential electrostatic impacts of histone tail acetylation. In collaboration with the Musselman Lab at the University of Colorado, we used our nucleosome technology to examine BPTF PHD-BD interactions2.
Using nucleosomes and histone peptides in parallel, we evaluated the BPTF PHD-BD binding. While the peptide data showed results consistent with previously published literature, the nucleosome data revealed strikingly different PTM binding preferences. Unlike on peptides, the BPTF PHD-BD did not engage with H4Kac nucleosomes, and showed >20-fold increased binding to combinatorial H3K4me3Kac over either H3 PTM alone. This synergistic effect was further characterized by NMR, which showed how the electrostatic impacts of H3Kac drive multivalent BPTF PHD-BD binding to the nucleosome – which had been missed in studies using histone peptides.
MLL1
MLL1, or mixed-lineage leukemia 1, encodes a highly conserved H3K4 methyltransferase that is frequently translocated/rearranged in acute lymphoid and myeloid leukemias. Interestingly, H3K4me3 almost always coincides with H3 lysine acetylation in vivo, but the precise mechanism underlying this combinatorial state remained unknown. In a 2023 collaboration with the Strahl lab at the University of North Carolina, we used our fully-defined nucleosome technology to investigate potential cross-talk between MLL1 activity, H3K4me3, and lysine acetylation3.
Specifically, we found that the MLL1 complex is more effective at generating H3K4me3 in the presence of H3 acetylation. We further dissected this mechanism using nucleosomes with H3K4 cis or trans to H3 acetylation, and found those with acetylation in cis to be the optimal substrates for MLL1 activity. The results support a mechanism in which lysine acetylation neutralizes electrostatic charge of the H3 tail, making H3 more accessible to MLL1 and thus supporting more efficient H3K4 methylation. Fully-defined nucleosomes were essential to these studies, as histone peptides completely missed this PTM interplay.
Other exciting applications
During the webinar, Dr. Keogh reviewed other exciting applications enabled by EpiCypher’s nucleosome substrates.
Epigenomic mapping with reader proteins
EpiCypher offers the ultra-sensitive epigenomic mapping assays, CUT&RUN and CUT&Tag, on our CUTANA™ platform. Traditionally, these assays use antibodies to localize the enzyme to the genomic target of interest. However, Dr. Keogh described work using a tagged recombinant BPTF PHD-BD as a detection reagent to uncover the genomic localization of combinatorial PTMs in cultured cells2. Importantly, EpiCypher was able to generate novel, fully-defined nucleosomes as physiological controls to monitor this assay and ensure confidence in the results.
Structural biology
Dr. Keogh highlighted the use of fully-defined nucleosomes to provide insights into structural biology. Cryogenic electron microscopy (cryo-EM) studies are key to understanding the specificity underlying chromatin interactions, and have been transformed by the availability of nucleosome substrates. To this end, he described recent cryo-EM research wherein our nucleosomes were used to uncover the structure of the Polycomb repressive deubiquitinase (PR-DUB) complex bound to a monoubiquitinated nucleosome (H2AK119Ub1). PR-DUB is a noted tumor suppressor and one of the most frequently mutated complexes in cancer, particularly in aggressive and/or early-onset cancers. Understanding how PR-DUB interacts with nucleosomes is critical to designing effective inhibitors and provides important insights into cancer etiology4.
Conclusions
Dr. Keogh demonstrated the importance of studying chromatin biology using a physiological substrate – the nucleosome. In particular, he provided insights into how EpiCypher scientists are driving advancements in chromatin biology and how our technologies are being used across the fields of biochemistry, genomics, and structural biology. Interested in learning more about these studies and the importance of nucleosomes in shaping the histone code? Watch Dr. Keogh’s talk on-demand now!
Stay tuned for more EpiCypher webinars! Something in particular you’d like to hear us talk about? Let us know!
References:
- Weinzapfel E.N. et al. Beyond the tail: the consequence of context in histone post-translational modification and chromatin research. Biochem J 481, 219-244 (2024). https://doi.org/10.1002/biot.201100155
- Marunde M. R. et al. Nucleosome conformation dictates the histone code. eLife 13, e78866. (2024). https://doi.org/10.7554/eLife.78866
- Jain K. et al. An acetylation-mediated chromatin switch governs H3K4 methylation read-write capability. eLife 12, e82596 (2023). https://doi.org/10.7554/eLife.82596
- Thomas J. F. et al. Structural basis of histone H2A lysine 119 deubiquitination by Polycomb repressive deubiquitinase BAP1/ASXL1. Sci Adv 9, eadg9832 (2023). https://doi.org/10.1126/sciadv.adg9832
