Starting CUT&RUN or CUT&Tag for a new target - What you need to know

CUT&RUN and CUT&Tag are ultra-sensitive chromatin mapping technologies gaining widespread popularity in the epigenomics field. These streamlined approaches are compatible with reduced cell inputs and lower sequencing depths compared to ChIP-seq, while simultaneously generating data with remarkable signal over background.
CUT&RUN or CUT&Tag?
EpiCypher scientists have optimized CUTANA™ CUT&RUN and CUT&Tag protocols for various targets, including histone post-translational modifications (PTMs) and transiently interacting chromatin-associated proteins such as transcription factors, chromatin reader proteins, and chromatin remodeling enzymes. Our most up-to-date CUT&RUN protocol is compatible with targets from each of the above categories and can generate robust data using as few as 5,000 cells (or nuclei) and 3-8 million sequencing reads.
The CUTANA Direct-to-PCR CUT&Tag protocol produces high quality histone PTM maps down to 1,000 cells (or nuclei) with only 3-8 million reads. Of note, our scientists have found CUT&Tag to be most reliable for histone PTMs as compared to chromatin-associated proteins. EpiCypher continues to recommend CUT&RUN as the go-to genomic mapping approach for all targets, while CUT&Tag is suggested for mapping histone PTMs from rare cell types or low starting cell numbers.
Native, Frozen, or Cross-linked – CUTANA™ protocols have you covered
Our CUT&RUN and CUT&Tag protocols have been adopted for a variety of cell processing methods, including native (non-fixed), cross-linked (fixed), and frozen cells or nuclei.
Download the CUTANA Cross-linking Protocol
As a result of these efforts, our researchers have gained valuable knowledge and defined a set of best practices for developing CUTANA assays to new histone PTMs (CUT&RUN or CUT&Tag) or chromatin associated proteins (CUT&RUN preferred). Surprisingly, we have found that moderate cross-linking of cells/nuclei is advantageous for certain targets. Read on to learn more!
When to consider cross-linking? Depends on your target
For the majority of targets, CUT&RUN performed under native conditions is the preferred workflow. We recommend testing antibodies in native CUT&RUN prior to using cross-linking conditions or CUT&Tag assays. For CUT&Tag workflows, native is also the best method.
However, there are situations where cross-linking is necessary to improve CUT&RUN or CUT&Tag signal. Below we outline a few examples of targets that may particularly benefit from light to moderate cross-linking (Table 1):
| Cell/nuclei preparation | Methods | Works well for |
|---|---|---|
| Native | No cross-linking | Majority of targets in CUT&RUN and CUT&Tag |
| Light to moderate cross-linking | 0.1 - 1% formaldehyde, 1 min |
Select targets in CUT&RUN, such as:
|
| Heavy cross-linking | 1% formaldehyde, 10 min | Standard ChIP conditions; NOT recommended for CUT&RUN |
However, it is nearly impossible to predict which targets will require cross-linking. Thus, for new or understudied targets, we recommend testing different strategies.
How to develop CUT&RUN or CUT&Tag assays to a new target
For researchers investigating a new target in CUT&RUN or CUT&Tag assays, EpiCypher scientists recommend a few key steps:
- Source three to five antibodies to the protein / histone PTM of interest, ideally targeting distinct epitopes. NOTE: Claims of “ChIP-grade” do not guarantee success in CUT&RUN or CUT&Tag! See this blog for more info.
- Always check for CUTANA CUT&RUN Antibodies to the target of interest, since these have been directly confirmed to work in CUT&RUN and come with native or cross-linking protocol recommendations.
- Test all of the antibodies in your CUT&RUN (or CUT&Tag) workflow, comparing cells (or nuclei) prepared under both native and lightly cross-linked conditions. Light cross-linking (0.1% formaldehyde, 1 min) is suggested as a starting point, since this generally preserves labile PTMs without negatively impacting DNA yields.
- We also recommend starting your tests with CUT&RUN assays, for the reasons outlined above. If the number of cells/nuclei is less than 5k, then CUT&Tag can be used. (histone PTMs preferred).
- The best cell prep conditions are selected based on a balance of DNA yields, enrichment, and signal-to-noise in the dataset (see examples below).
- If light-cross linking and native conditions don't generate quality data, moderate cross-linking (1% formaldehyde, 1 min) may also be used.
In the rest of this blog, we will cover examples of how light to moderate cross-linking can improve CUT&RUN signal for lysine acetylation PTMs and acetyl-binding reader proteins.
Cross-linking in CUT&RUN to histone PTMs
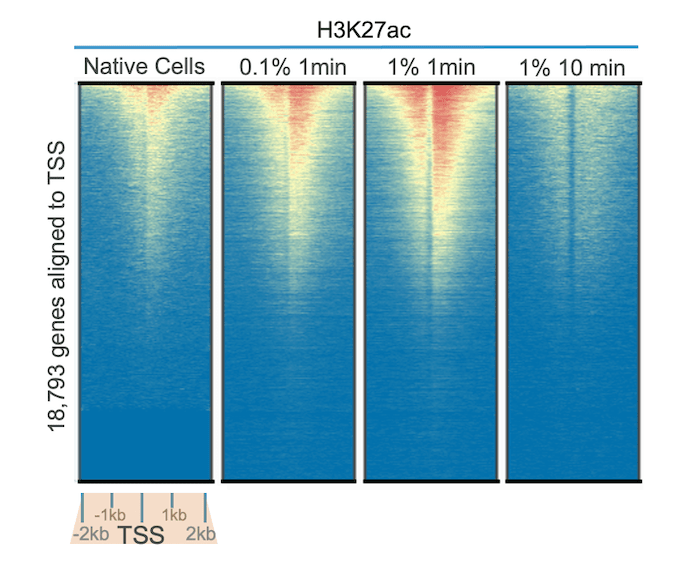
In Figure 1, we tested our highly specific H3K27ac antibody (EpiCypher 13-0045) in CUT&RUN assays, using native cells and cells treated with varying concentrations of formaldehyde for 1 to 10 minutes. Genome-wide heatmap analysis reveals improved signal over background with 0.1% and 1% formaldehyde treatment for 1 minute compared to no cross-linking. This is partly explained by the high lability of many acetyl marks, as acetylation is known to be dynamically regulated and display increased turnover relative to other classes of PTMs (e.g. methylation)1.
Interestingly, these results also show that over-fixation in CUT&RUN can reduce signal and recovery. Treatment with 1% formaldehyde for 10 minutes reduced yield and diminished signal over background compared to all other conditions tested, including non-fixed cells. These results are notable, as most ChIP-seq protocols recommend 1% formaldehyde cross-linking for 10 minutes. Thus, for acetylation PTMs, we highly recommend testing samples with light to moderate cross-linking protocols to investigate benefits for your assay.
Notably, while
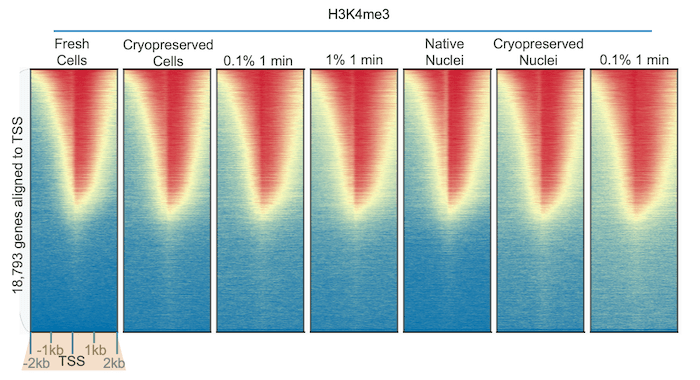
some PTMs are altered by fixation, others are unaffected. We used our
robust H3K4me3 antibody (EpiCypher 13-0041) to perform CUT&RUN using K562 cells subjected to diverse preparation
methods. The results were identical across all samples (Figure 2),
illustrating the stability of histone methylation1 and the
reliability of our CUT&RUN protocol.
Cross-linking in CUT&RUN to chromatin-associated proteins
Importantly, CUT&RUN for non-histone PTM targets can also benefit from light to moderate fixation. Our recently released antibodies to chromatin remodeling proteins BRM (SMARCA2) and SNF2L (SMARCA1) both display enhanced recovery and peak amplitude when using cross-linked vs. native samples.
Figure 3 shows representative CUT&RUN data for our BRM antibody, including heatmaps and loci-specific peak analysis at transcription start sites (TSSs). H3K4me3 data are shown for comparison, as chromatin remodelers typically function to rearrange nucleosomes at TSSs and are often correlated with H3K4me3 levels2. Fixation with 0.1% formaldehyde for 1 minute vastly improved signal : noise at TSSs, more closely replicating H3K4me3 peak patterns (Figure 3A) vs. non-fixed cells. These results are even more apparent when looking at BRM target loci, such as the transcription factor POU2F1 and microtubule-associated protein MAP4. In Figure 3B, genomic mapping data for native vs. cross-linked CUT&RUN reactions at these loci are outlined using red boxes. The native reactions display peaks barely discernable over background. In contrast, CUT&RUN data from moderately cross-linked cells exhibit increased peak amplitude, while maintaining extremely low backgrounds characteristic of CUT&RUN.
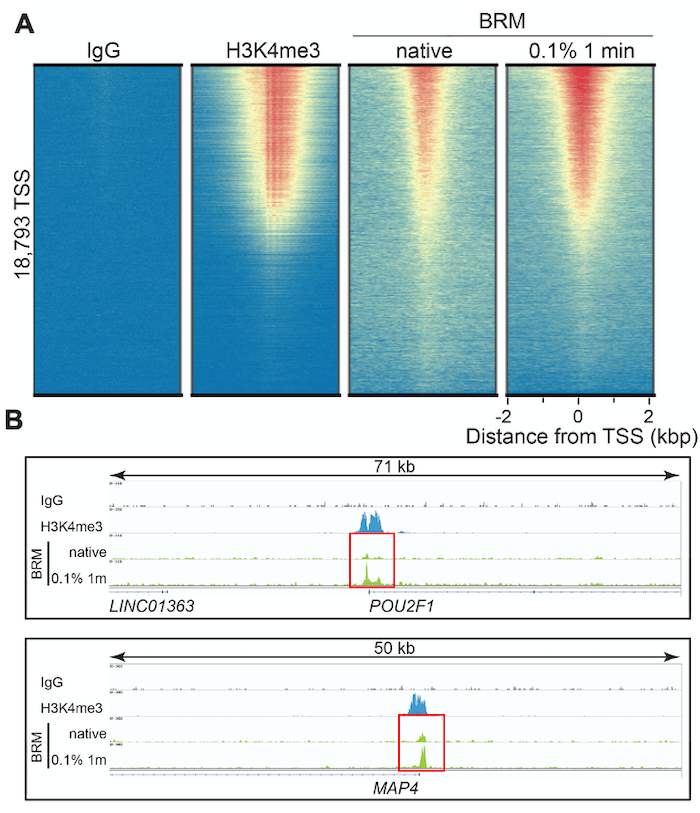
It is not clear how or why fixation augments recovery for some of these chromatin-associated proteins. In the case of BRM, it may be due to the presence of a C-terminal bromodomain in the BRM protein. Bromodomains interact with acetylated histones; thus, fixation may stabilize lysine acetylation – BRM interactions, enabling improved detection by CUT&RUN.
Get the CUTANA CUT&RUN Protocol
Conclusions: Test your antibody
The overarching summary of EpiCypher’s extensive research efforts is that before embarking on a new CUT&RUN or CUT&Tag project, you should test a variety of conditions (antibodies, preparation methods) to find the optimal methods for your target of interest. This will accelerate your future studies by developing a robust workflow, thereby reducing frustrating troubleshooting steps and increasing confidence in your data.
References
- Zee BM et al. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin 3, 22 (2010). PubMed PMID: 21134274.
- Wysocka J et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86-90 (2006). PubMed PMID: 16728976.
