SPRI bead or column DNA purification: Which method is best for CUT&RUN?

Purification of target-enriched DNA is central to CUT&RUN (and CUT&Tag) chromatin mapping workflows. When considering a DNA cleanup strategy, multiple methods are available: phenol-chloroform extraction, column-based separation, and SPRI bead technology. With so many options, how can you be sure you’ve selected the right method? EpiCypher is here to help. We are a global leader in CUT&RUN assay development, and many of our protocol modifications have become standard in the industry. As a result of these efforts, the newest version of the CUTANA™ CUT&RUN Kit leverages SPRI beads for DNA purification, replacing older column formats.
Start your experiment now with the CUTANA™ CUT&RUN Kit
In this blog we will address several questions and highlight the utility of SPRI beads for DNA purification in CUT&RUN.
- Why did we remove spin columns from the CUTANA™ CUT&RUN Kit?
- How does SPRI bead DNA purification work?
- What are the advantages of SPRI beads for CUT&RUN? Why should I consider making the switch?
- Are CUT&RUN yields the same when comparing column- and bead-based DNA purification strategies?
- Why are SPRI beads better for size selection of sequencing libraries?
- What are the most frequent concerns with SPRI beads, and where do I go for help?
Why did we remove spin columns from the CUTANA™ CUT&RUN Kit?
The main factors that led to removal of DNA purification spin columns from the CUTANA™ CUT&RUN Kit relate to experimental reproducibility and throughput. Assay reliability has been a major focus of EpiCypher R&D since we first started working on CUT&RUN in 2019. Our initial efforts included optimizing the CUT&RUN workflow for 8-strip tubes and multi-channel pipettors, which reduced hands-on time and minimized sample variation. The use of 8-strip tubes, as opposed to 1.5 mL tubes, also allowed our scientists to process more CUT&RUN reactions in a single experiment.
The enhanced reproducibility of the CUTANA™ CUT&RUN protocol opened the door to epigenomics applications, such as clinical research and biomarker development. The low cell number requirements also meant that scientists could map more targets, include additional replicates, analyze diverse sample conditions, and run more controls. These advantages set the stage for automated CUT&RUN assays – with one minor holdup: the use of spin columns for DNA purification.
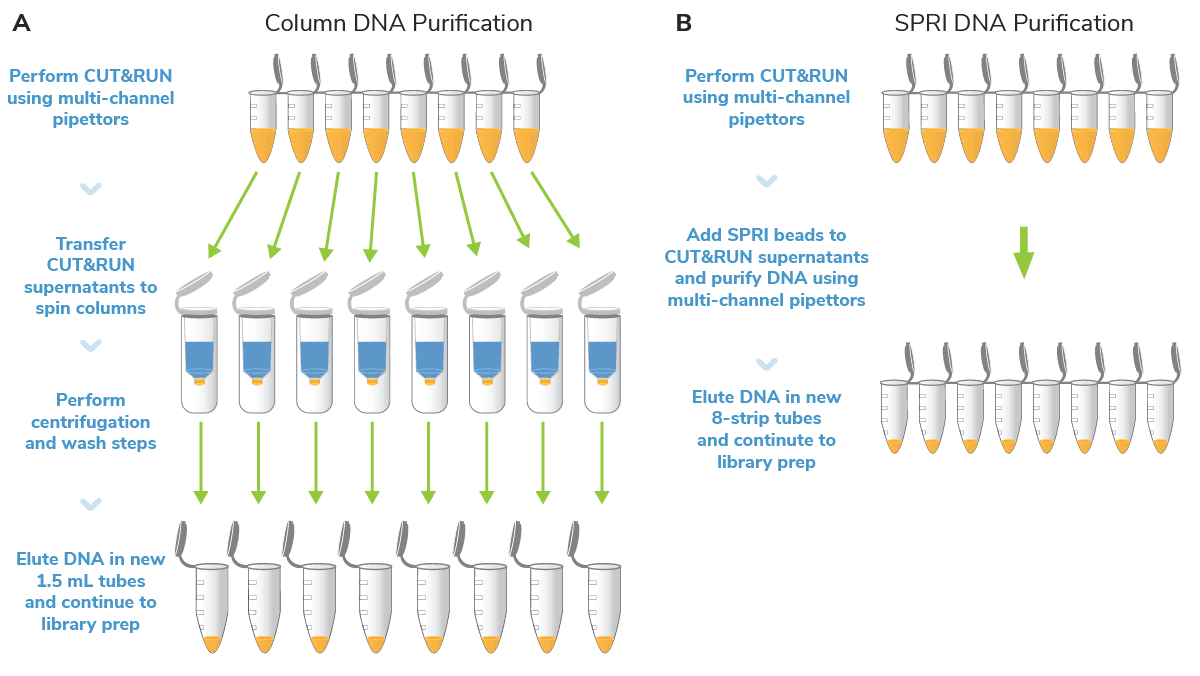
In our original CUT&RUN workflow, users transfer supernatants containing targeted-enriched DNA from 8-strip tubes to 1.5 mL tubes, and then perform a standard column-based cleanup. Each sample must be individually and manually processed, which limits the number of samples that can be run in a single experiment and negatively impacts reproducibility (Figure 1A). It also adds significant time to the protocol. For instance, EpiCypher scientists routinely perform CUT&RUN experiments with 48+ reactions, which is straightforward with 8-strip tubes and multi-channel pipettors. But processing 48+ spin columns for DNA cleanup is a daunting task, even for our seasoned bench scientists!
In searching for robust DNA purification methods compatible with 8-strip tubes, we quickly found a standout candidate in SPRI beads. We already used SPRI beads for our CUTANA™ CUT&Tag Kit, and after adopting SPRI bead-based DNA cleanup for our automated CUTANA™ CUT&RUN Services, our scientists quickly realized their potential for CUT&RUN kits. With SPRI beads, scientists can now go from cells to final purified CUT&RUN DNA in 8-strip tubes, saving precious time and improving assay reliability (Figure 1B). SPRI beads also provide higher-quality DNA – keep reading to learn more.
How does SPRI bead DNA purification work?
SPRI technology (Solid-Phase Reversible Immobilization) uses a combination of magnetic fields and ionic charge to purify DNA. In this method, negatively charged paramagnetic beads are prepared in a buffer containing the “crowding reagent” polyethylene glycol (PEG) and high salt.
Beads are then added to DNA at a specific volumetric ratio, which alters the charge of the solution and the concentration of PEG. These changes to pH impact the size of DNA fragments that bind the beads. Higher bead:DNA ratios capture DNA fragments of varying lengths, while lower ratios preferentially recover longer fragments.
A magnetic rack is used to collect beads, and the supernatant is discarded. Beads are washed with 85% ethanol to remove impurities. DNA is then released, or eluted, from beads using a physiological, low salt buffer. This eluted material can be utilized in downstream applications, including sequencing library preparation.
What are the advantages of SPRI beads for CUT&RUN? Why should I consider making the switch?
There are numerous advantages to using SPRI beads over spin columns for CUT&RUN DNA purification. Though columns are a mainstay of DNA purification kits and have been used for decades in molecular biology, SPRI beads are preferrable in many genomics applications.
The main advantage of using magnetic SPRI beads vs. columns is the reduced number of sample processing steps (Figure 1), which makes the entire CUT&RUN workflow more streamlined. These features result in several downstream benefits:
- Faster, more robust DNA purification workflow. There are no individual column transfers or centrifugation steps, thus reducing the overall protocol time (Figure 1). Fewer protocol steps help minimize risk of sample loss, DNA shearing, or other human errors that can negatively impact reproducibility.
- Greater experimental throughput and reliability. The CUT&RUN workflow is fully compatible with 8-strip tubes, magnets, and multi-channel pipettors to reduce sample handling variation and enable scaling for large projects.
- Improved size-selection for sequencing libraries. Magnetic beads provide broad flexibility for various applications in genomics, including sequencing library size selection and cleanup. We provide a detailed protocol for SPRI bead sequencing library size selection in the CUTANA™ Quick Cleanup DNA Purification Kit manual.
Are CUT&RUN yields the same when comparing column- and bead-based DNA purification strategies?
Yes, CUT&RUN yields across the column and SPRI bead DNA are roughly the same. As part of our CUTANA™ CUT&RUN Kit update, our product development team rigorously compared beads vs. columns in our CUT&RUN workflow, testing multiple targets, cell numbers, and native vs. cross-linking conditions (Figure 2A-B).
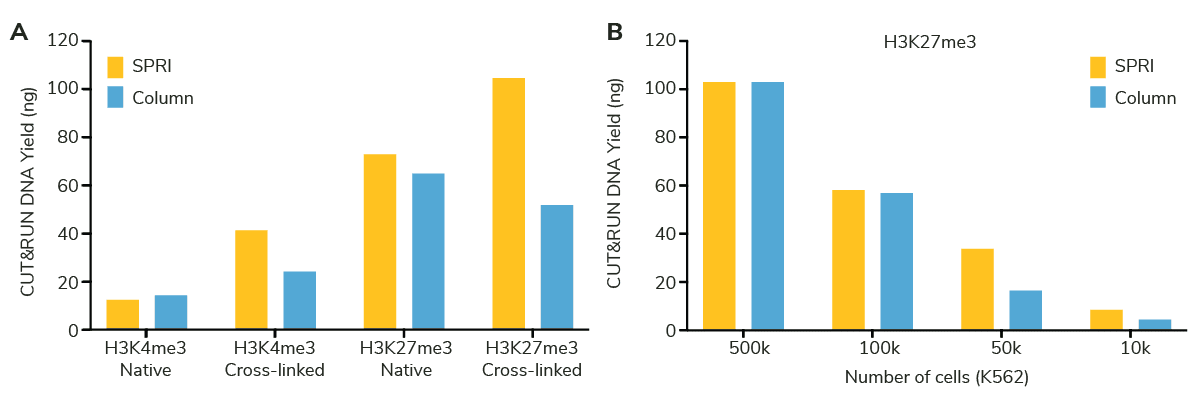
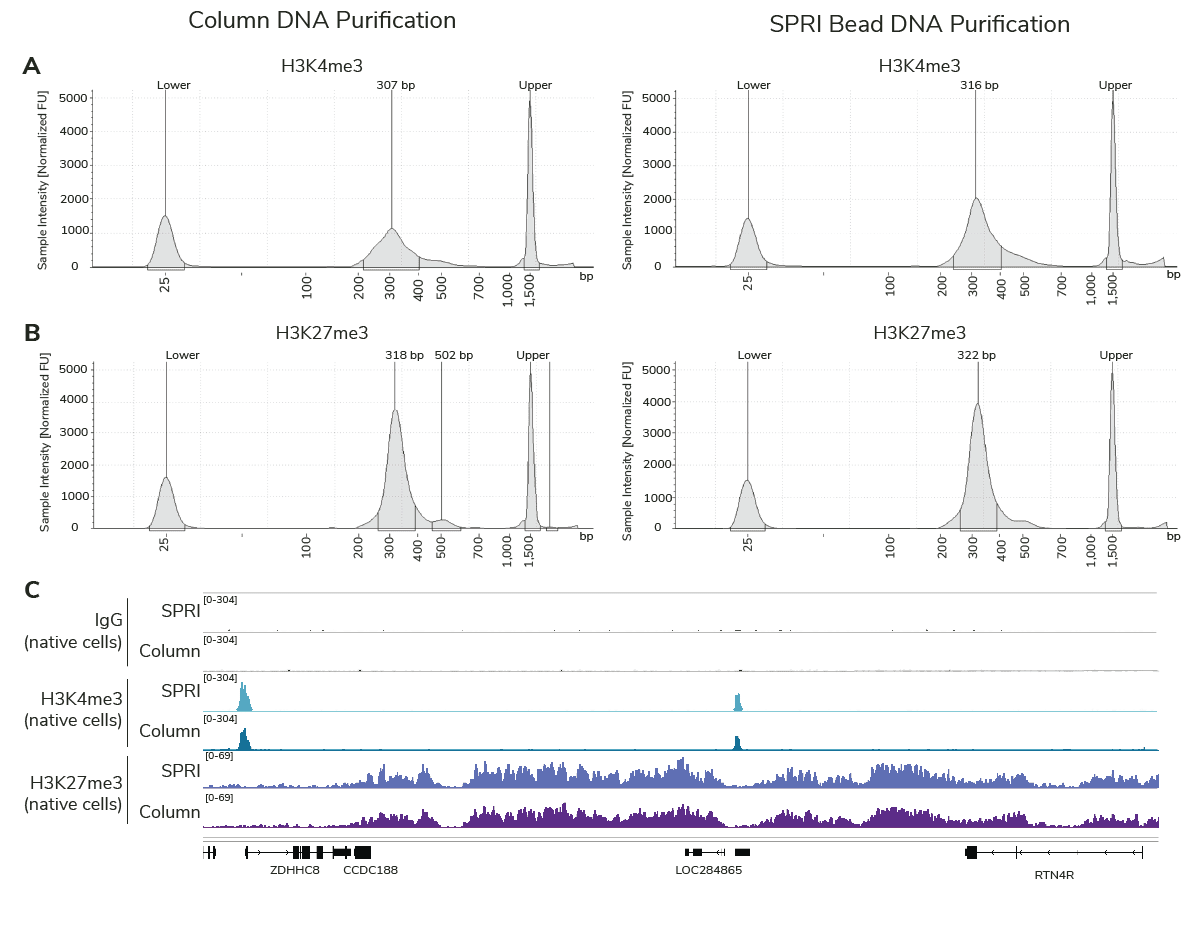
Importantly, the fragment distribution traces of purified sequencing libraires – our recommended metric for CUT&RUN assay success – also show appropriate size selection using SPRI beads (Figure 3A-B) and nearly indistinguishable sequencing data (Figure 3C). Together, these experiments show that the two DNA cleanup methods generate comparable (in some cases, better!) DNA yields and downstream sequencing data.
Why are SPRI beads better for size selection of sequencing libraries?
SPRI beads provide a more sensitive method for library size selection and cleanup with increased DNA recovery compared to older gel extraction methods. In CUT&RUN, size selection is necessary when sequencing libraries contain a high percentage of adapter dimers, which appear at ~150 bp in Bioanalyzer or TapeStation fragment distribution traces.
To remove adapter dimers, many kits recommend a gel purification approach, in which pooled sequencing libraries are run on an agarose gel containing a DNA intercalating dye (e.g. ethidium bromide). The ~300 bp library band is carefully – but quickly – excised under UV light using a small razor blade and purified using labor-intense centrifugation and wash steps. This manual, low-tech solution comes with significant safety concerns and typically results in ~70% library loss. Generating high-quality sequencing data with adequate coverage can be challenging in these cases.
At EpiCypher, we utilize SPRI beads for size selection of pooled CUT&RUN libraries, as outlined in the CUTANA™ Quick Cleanup DNA Purification Kit manual. The use of SPRI technology provides a major upgrade over gel purification, allowing you to concentrate pooled libraries and remove unwanted short fragments in one standardized, easy workflow. DNA damage and/or loss is minimized, as everything is performed on magnetic beads; no gel electrophoresis or UV light is needed.
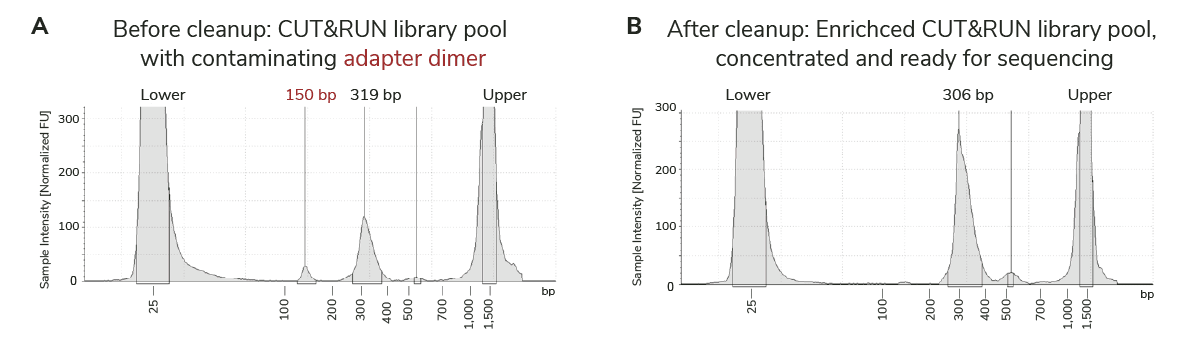
Figure 4 shows TapeStation traces from a CUT&RUN library pool before and after removal of adapter dimers using the CUTANA™ Quick Cleanup DNA Purification Kit. In this Figure, we see that the unwanted smaller fragments (adapter dimers) are preferentially removed, while the larger library fragments are retained – a highly specific result made possible by SPRI beads.
What are the most frequent concerns with SPRI beads, and where do I go for help?
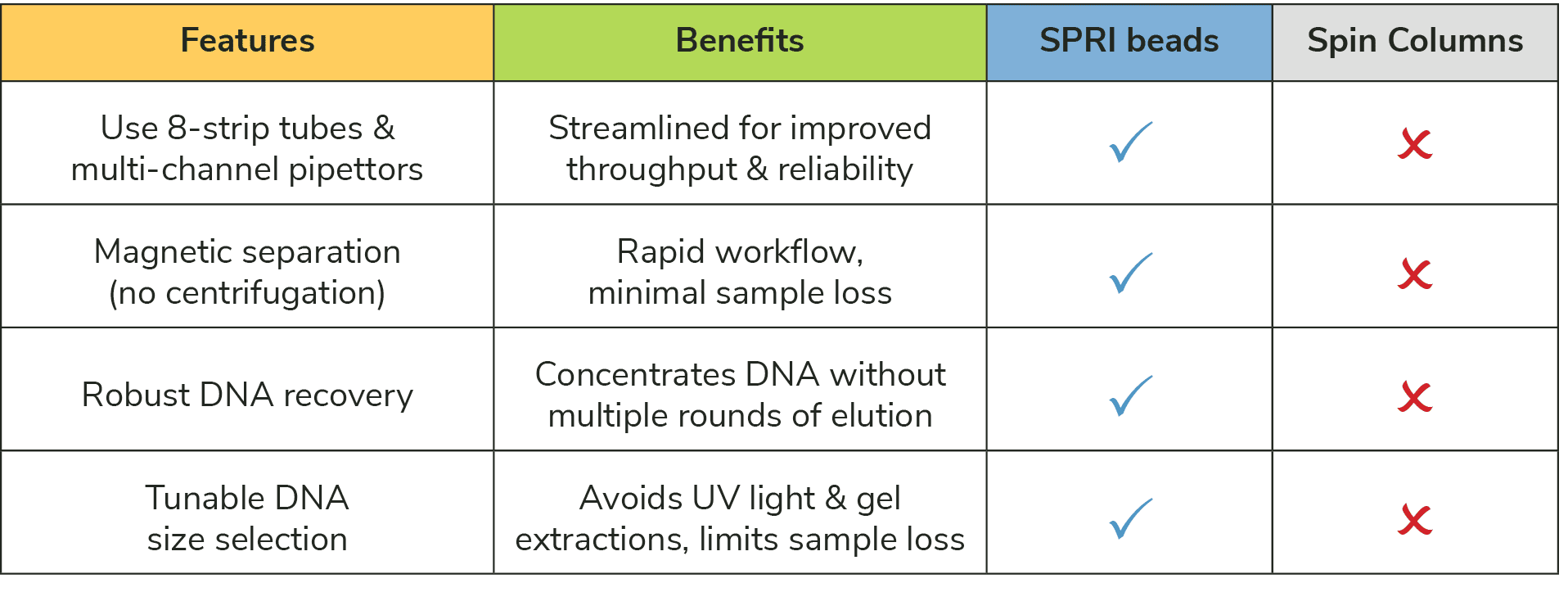
Switching to bead-based DNA purification can be intimidating and frustrating, especially when you are at the final steps of an important assay. EpiCypher’s Tech Support Team is here to help you make the transition from spin columns to SPRI and can answer any technical questions you might have. Ultimately, this CUT&RUN kit update will help generate more reproducible data, save time and frustration, and support larger experimental designs (Table 1). Our scientists routinely run 30-50 reactions per experiment!
For the updated CUT&RUN protocol, check out our latest CUTANA™ CUT&RUN Kit Manual here, or visit EpiCypher’s Tech Support Center. Reach out to techsupport@epicypher.com with any questions.
