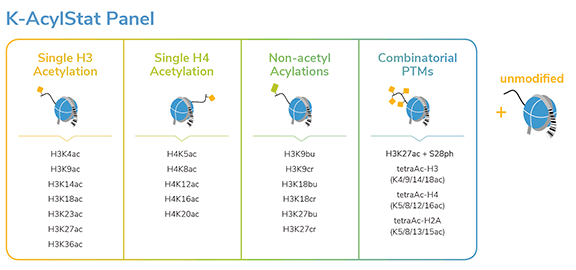
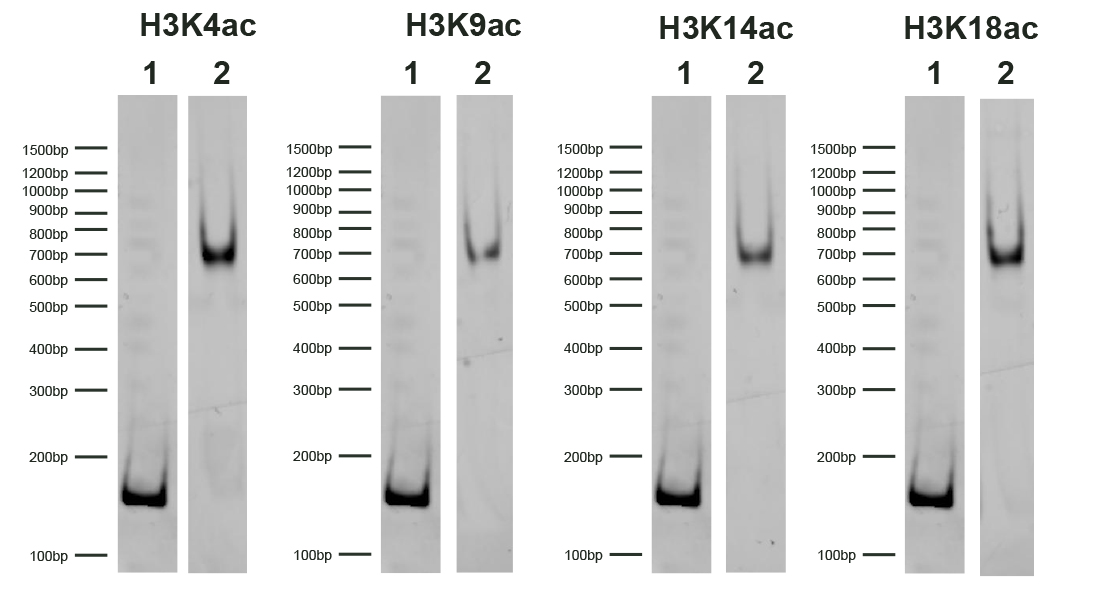
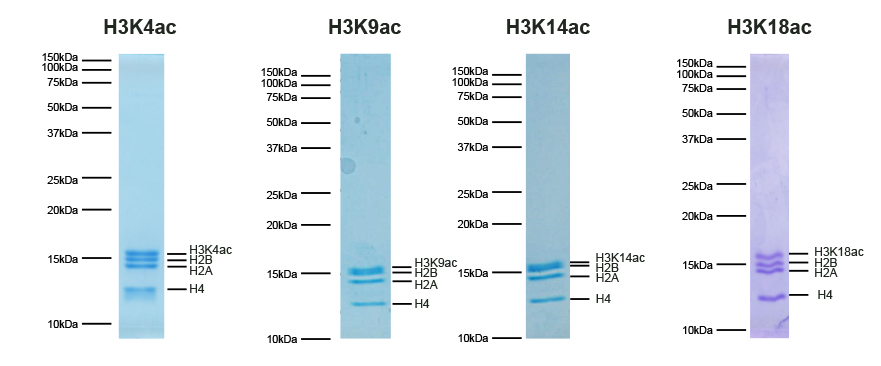
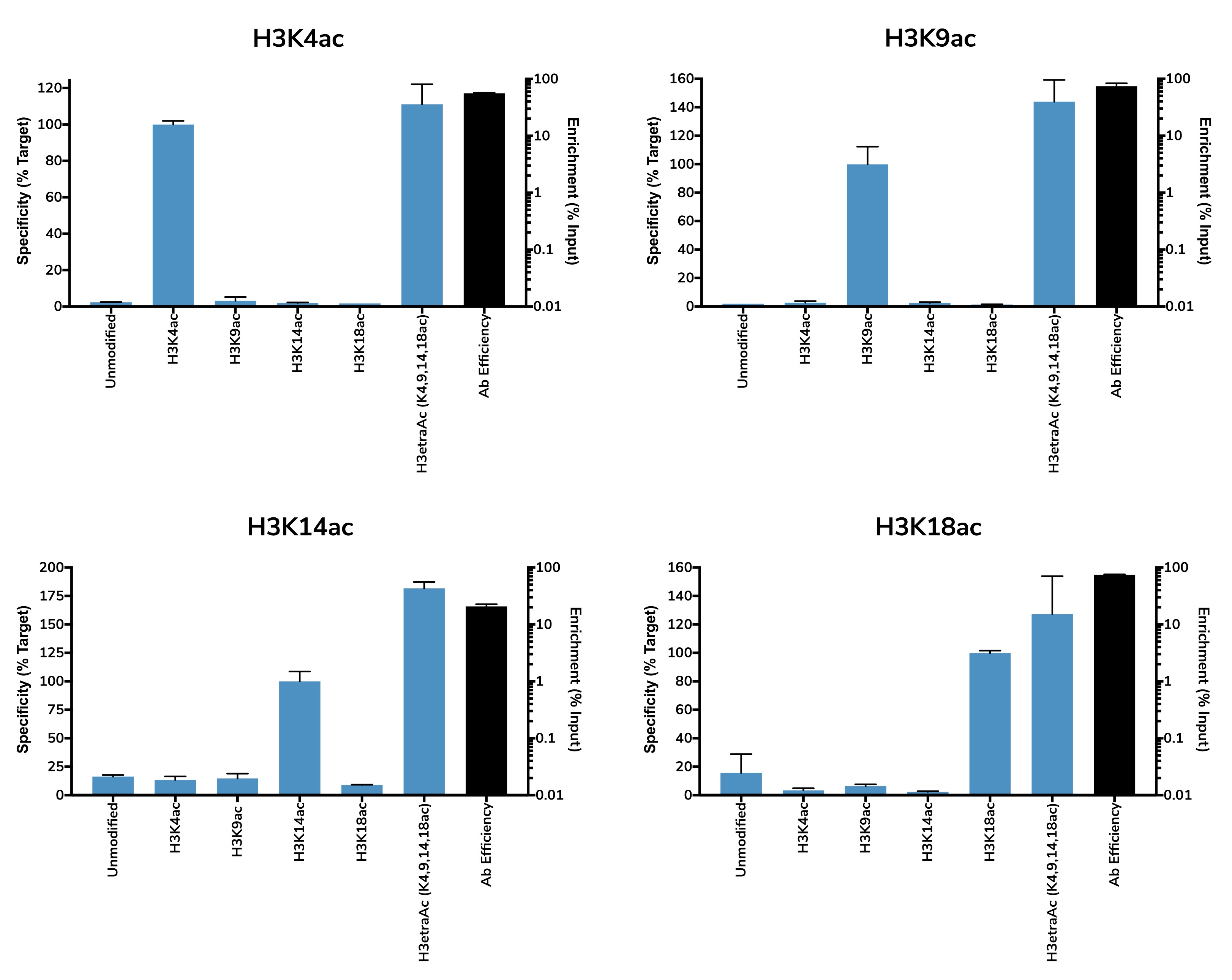
SNAP-ChIP® K-AcylStat Panel
$365.00
SKU: 19-3001
{"sku":"19-3001","url":"https://www.epicypher.com/products/nucleosomes/snap-chip-k-acylstat-panel","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"options":[{"id":704,"type":"Configurable_PickList_Set","display_name":"Select Pack Size:","required":true,"condition":true,"state":"variant_option","values":[{"label":"20 µl","id":111,"data":"20 µl","selected":true},{"label":"200 µl","id":106,"data":"200 µl","selected":false}],"partial":"set-radio"}],"id":687,"bulk_discount_rates":[],"can_purchase":true,"meta_description":"SNAP-ChIP K-AcylStat Panel Spike-in Normalization Control for Chromatin IP (ChIP) and ChIP-Seq","category":["Nucleosomes","Nucleosomes/SNAP-ChIP<sup>®</sup> Spike-ins"],"main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/687/654/SNAP_Chip__17660.1557847064.png?c=2","alt":"SNAP-ChIP® K-AcylStat Panel"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=687","shipping":{"calculated":true},"num_reviews":0,"weight":"0.01 LBS","description":"<table border=\"0\" cellpadding=\"2\" cellspacing=\"2\" width=\"100%\">\n\t<tbody>\n\t\t<tr valign=\"top\">\n\t\t\t<td>\n\t\t\t<table border=\"0\" cellpadding=\"1\" cellspacing=\"1\" height=\"1490\" width=\"90%\">\n\t\t\t\t<tbody>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td valign=\"top\">\n\t\t\t\t\t\t<p></p>\n\n\t\t\t\t\t\t\n\t\t\t\t\t\t<span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b><b>SNAP-ChIP K-AcylStat Panel</b> Description:</b> A panel of distinctly modified mononucleosomes assembled from recombinant human histones expressed in E. coli (two each of histones H2A, H2B, H3 and H4; accession numbers: H2A-P04908; H2B-O60814; H3.1-P68431 or H3.2-Q71DI3*; H4-P62805) wrapped by 147 base pairs of barcoded Widom 601 positioning sequence DNA. The panel is comprised of a pool of 1 unmodified plus 22 histone H2A, H3, or H4 post-translational modifications (PTMs, created by a proprietary semi-synthetic method): H3K4ac, H3K9ac, H3K14ac, H3K18ac, H3K23ac, H3K27ac, H3K36ac, H4K5ac, H4K8ac, H4K12ac, H4K16ac, H4K20ac, H3K9bu, H3K9cr, H3K18bu, H3K18cr, H3K27bu, H3K27cr, H3K27acS28phos, H3K4,9,14,18ac, H4K5,8,12,16ac, H2AK5,8,13,15ac. Each distinctly modified nucleosome is distinguishable by a unique sequence of DNA (“barcode”) at the 3’ end that can be deciphered by qPCR or next-generation sequencing. Each of the 23 nucleosomes in the pool is wrapped by 2 distinct DNA species, each containing a distinct barcode (”A” and “B”, see SNAP-ChIP Manual) allowing for an internal technical replicate.</span></span>\n\n\t\t\t\t\t\t<p><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><span style=\"color: orange !important;\"><b>Learn more about SNAP-ChIP Certified Antibodies <a href=\"https://www.epicypher.com/snap-chip-certified-antibodies/\">here</a></b></span></span></span></p>\n\n\t\t\t\t\t\t<p><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\">* Histone H3.2 contains a Cys to Ala mutation at position 110. </span></span><br />\n\t\t\t\t\t\t</p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td valign=\"top\"><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b><b>SNAP-ChIP K-AcylStat Panel</b> Formulation:</b> PPurified recombinant mononucleosomes, containing a mixture of 23 (1 unmodified plus 22 unique) H2A, H3, and H4 PTMs in 10 mM sodium cacodylate, pH 7.5, 100 mM NaCl, 1 mM EDTA, 50% glycerol (w/v), 1x Protease Inhibitor cocktail, 100 µg/mL BSA, 10 mM β-mercaptoethanol. Average molarity = 0.6 nM. MW = ~199382.1 Da (average MW of all 23 nucleosomes).</span></span><br /><br />\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td valign=\"top\"><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b>SNAP-ChIP K-AcylStat Panel Storage and Stability:</b> Stable for six months at -20°C from date of receipt.</span></span><br /><br />\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td valign=\"top\"><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b>SNAP-ChIP K-AcylStat Panel Application Notes:</b> SNAP-ChIP K-AcylStat barcoded nucleosome standards are highly purified recombinant mononucleosomes and are suitable for use as spike-in controls for ChIP reactions, for antibody specificity testing or for effector protein binding experiments. See product manual for more information. SNAP-ChIP can easily be introduced into existing ChIP and ChIP-seq workflows for experiment normalization and monitoring of technical variability. </span></span><br /><br />\n\t\t\t\t\t\t<p><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><span style=\"color: orange !important;\"><b>Request a quote <a href=\"https://www.epicypher.com/request-a-quote/\">here</a></b></span></span></span></p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td valign=\"top\">\n\t\t\t\t\t\t<p><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b>Background References</b>:<br />\n\t\t\t\t\t\tGrzybowski A et al. 2015 Mol Cell 58: 886 - 899 <a href=\"https://www.ncbi.nlm.nih.gov/pubmed/26004229\" target=\"_new\" title=\"Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide\">LINK</a></span></span></p>\n\n\t\t\t\t\t\t<p><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b>Product References</b>:<br />\n\t\t\t\t\t\tTay RE et al. 2020 J Exp. Med. 217: e20191453 <a href=\"https://www.ncbi.nlm.nih.gov/pubmed/32374402\" target=\"_new\" title=\"Hdac3 is an epigenetic inhibitor of the cytotoxicity program in CD8 T cells\">LINK</a></span></span><br />\n\t\t\t\t\t\t</p>\n\n\t\t\t\t\t\t<p>\n\t\t\t\t\t\t</p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td valign=\"top\">\n\t\t\t\t\t\t<p><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b>SNAP-ChIP K-AcylStat Panel Additional Resources and Files:</b></span></span></p>\n\n\t\t\t\t\t\t<table border=\"0\" cellpadding=\"0\" cellspacing=\"10\" height=\"447\" width=\"450\">\n\t\t\t\t\t\t\t<tbody>\n\t\t\t\t\t\t\t\t<tr>\n\t\t\t\t\t\t\t\t\t<td style=\"height:60px\" valign=\"top\" width=\"350\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>Technical Datasheet </strong></span></span></td>\n\t\t\t\t\t\t\t\t\t<td align=\"center\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/documents/tds/19-3001.pdf\" target=\"_new\"><img alt=\"19-3001 Datasheet\" height=\"40\" src=\"/content/documents/tds/icon.png\" width=\"30\" /></a></span></span></td>\n\t\t\t\t\t\t\t\t</tr>\n\t\t\t\t\t\t\t\t<tr>\n\t\t\t\t\t\t\t\t\t<td style=\"height:60px\" valign=\"top\" width=\"350\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>SNAP-ChIP K-AcylStat Manual</strong></span></span></td>\n\t\t\t\t\t\t\t\t\t<td align=\"center\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/documents/snapchip/SNAP-ChIP_K-AcylStat_Manual.pdf\" target=\"_new\"><img alt=\"SNAP-ChIP K-AcylStat manual.pdf\" height=\"40\" src=\"/content/documents/tds/icon.png\" width=\"30\" /></a></span></span></td>\n\t\t\t\t\t\t\t\t</tr>\n\t\t\t\t\t\t\t\t<tr>\n\t\t\t\t\t\t\t\t\t<td style=\"height:60px\" valign=\"top\" width=\"350\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>SNAP-ChIP Brochure</strong></span></span></td>\n\t\t\t\t\t\t\t\t\t<td align=\"center\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/docs/SNAP-ChIP.pdf\" target=\"_new\"><img alt=\"SNAP-ChIP K-AcylStat manual.pdf\" height=\"40\" src=\"/content/documents/tds/icon.png\" width=\"30\" /></a></span></span></td>\n\t\t\t\t\t\t\t\t</tr>\n\t\t\t\t\t\t\t\t<tr>\n\t\t\t\t\t\t\t\t\t<td style=\"height:60px\" valign=\"top\" width=\"350\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>SNAP-ChIP K-AcylStat Full Panel Information: </strong>Forward/reverse barcodes, nucleosomal DNA sequences, qPCR primer sequences, TaqMan probe sequence</span></span></td>\n\t\t\t\t\t\t\t\t\t<td align=\"center\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/documents/snapchip/SNAP-ChIP_K-AcylStat_19-3001_Barcode-Sequence-Primer_Info-SHORT_ver3.0.xlsx\" target=\"_new\"><img alt=\"19-3001 Datasheet\" height=\"40\" src=\"/content/documents/excelicon.png\" width=\"30\" /></a></span></span></td>\n\t\t\t\t\t\t\t\t</tr>\n\t\t\t\t\t\t\t\t<tr>\n\t\t\t\t\t\t\t\t\t<td style=\"height:60px\" valign=\"top\" width=\"350\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>SNAP-ChIP K-AcylStat qPCR Primer Sequences</strong> - Formatted for easy upload for ordering from common vendors (<em>e.g.</em> IDT)</span></span></td>\n\t\t\t\t\t\t\t\t\t<td align=\"center\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/documents/snapchip/SNAP-ChIP_K-AcylStat_qPCR_Primers_BarcodeA_v2.0.xlsx\" target=\"_new\"><img alt=\"19-3001 Datasheet\" height=\"40\" src=\"/content/documents/excelicon.png\" width=\"30\" /></a></span></span></td>\n\t\t\t\t\t\t\t\t</tr>\n\t\t\t\t\t\t\t\t<tr>\n\t\t\t\t\t\t\t\t\t<td style=\"height:60px\" valign=\"top\" width=\"350\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>Fasta formatted file of K-AcylStat barcode Sequences</strong> - For alignment of NGS sequencing data</span></span></td>\n\t\t\t\t\t\t\t\t\t<td align=\"center\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/documents/snapchip/KacylStat_barcoded_fasta_v2.0.rtf\" target=\"_new\"><img alt=\"19-3001 fasta\" height=\"40\" src=\"/content/documents/excelicon.png\" width=\"30\" /></a></span></span></td>\n\t\t\t\t\t\t\t\t</tr>\n\t\t\t\t\t\t\t</tbody>\n\t\t\t\t\t\t</table>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t</tbody>\n\t\t\t</table>\n\t\t\t</td>\n\t\t\t<td>\n\t\t\t<table border=\"0\" cellpadding=\"3\" cellspacing=\"3\" width=\"100%\">\n\t\t\t\t<tbody>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"center\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/images/nucleosome/k-acylstat_panel.gif\" target=\"_new\"><img alt=\"K-AcylStat Panel\" src=\"/content/images/nucleosome/k-acylstat_panel.gif\" width=\"250\" /></a></span></span></td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"left\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>SNAP-ChIP K-AcylStat Panel:</strong> Schematic representation of nucleosomes comprising the K-AcylStat Panel: H3K4ac, H3K9ac, H3K14ac, H3K18ac, H3K23ac, H3K27ac, H3K36ac, H4K5ac, H4K8ac, H4K12ac, H4K16ac, H4K20ac, H3K9bu, H3K9cr, H3K18bu, H3K18cr, H3K27bu, H3K27cr, H3K27acS28phos, H3K4,9,14,18ac, H4K5,8,12,16ac and unmodified.<br />\n\t\t\t\t\t\t<strong>(Click image to enlarge) </strong></span></span>\n\t\t\t\t\t\t<p></p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t</tbody>\n\t\t\t</table>\n\n\t\t\t<table border=\"0\" cellpadding=\"3\" cellspacing=\"3\" width=\"100%\">\n\t\t\t\t<tbody>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"center\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/images/products/nucleosomes/19-3001-DNA-gel.jpg\" target=\"_new\"><img alt=\"19-3001 DNA Gel Data\" src=\"/content/images/products/nucleosomes/19-3001-DNA-gel.jpg\" width=\"250\" /></a></span></span></td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"left\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>DNA Gel Data:</strong> Representative images for SNAP-ChIP K-AcylStats resolved by native PAGE and stained with ethidium bromide to visualize DNA. Lane 1: Free 147bp DNA used in nuclesome assembly (100 ng). Lane 2: Intact nucleosomes (200 ng). Comparable experiments were performed for the entire K-AcylStat Panel.<br />\n\t\t\t\t\t\t<strong>(Click image to enlarge) </strong></span></span>\n\t\t\t\t\t\t<p></p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t</tbody>\n\t\t\t</table>\n\n\t\t\t<table border=\"0\" cellpadding=\"3\" cellspacing=\"3\" width=\"100%\">\n\t\t\t\t<tbody>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"center\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/images/products/nucleosomes/19-3001-protein-gel.jpg\" target=\"_new\"><img alt=\"19-3001 Protein Gel Data\" src=\"/content/images/products/nucleosomes/19-3001-protein-gel.jpg\" width=\"250\" /></a></span></span></td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"left\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>Protein Gel Data:</strong> Representative Coomassie stained PAGE gel of SNAP-ChIP K-AcylStat (2 µg each) to demonstrate the purity of the histones in the preparation. Sizes of molecular weight markers and positions of the core histones (H2A, H2B, H3 and H4) are indicated. Comparable experiments were performed for the entire K-AcylStat Panel.<br />\n\t\t\t\t\t\t<strong>(Click image to enlarge) </strong></span></span>\n\t\t\t\t\t\t<p></p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t</tbody>\n\t\t\t</table>\n\n\t\t\t<table border=\"0\" cellpadding=\"3\" cellspacing=\"3\" width=\"100%\">\n\t\t\t\t<tbody>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"center\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/images/products/nucleosomes/19-3001-ChIP-data.jpg\" target=\"_new\"><img alt=\"19-3001 ChIP Data\" src=\"/content/images/products/nucleosomes/19-3001-ChIP-data.jpg\" width=\"250\" /></a></span></span></td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"left\" valign=\"top\">\n\t\t\t\t\t\t<p><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>ChIP Data:</strong> Representative chromatin imunoprecipitation (ChIP) data using commerically available ChIP-grade antibodies targeting the indicated PTMs. The antibodies were assayed in a native ChIP experiment with 3 μg antibody added to 3 μg K-562 chromatin with the K-AcylStat Panel spiked-in prior to micrococcal nuclease digestion. Quantitative real-time PCR (qPCR) was used to measure recovery of duplicate DNA barcodes corresponding to each uniquely modified nucleosome in the panel (blue bars, X-axis). The black bars map to the log scale on the right y-axis and indicate the percentage of target immunoprecipitated relative to the input (a measure of the antibody efficiency). In each case, the SNAP-ChIP spike-in confirmed that the antibodies recovered the expected histone PTM with high efficiency and specificity. Comparable experiments were performed for the entire K-AcylStat Panel.<br />\n\t\t\t\t\t\t<strong>(Click image to enlarge) </strong></span></span></p>\n\n\t\t\t\t\t\t<p></p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t</tbody>\n\t\t\t</table>\n\n\t\t\t<table border=\"0\" cellpadding=\"3\" cellspacing=\"3\" width=\"100%\">\n\t\t\t\t<tbody>\n\t\t\t\t</tbody>\n\t\t\t</table>\n\t\t\t</td>\n\t\t</tr>\n\t</tbody>\n</table>\n\n","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$365.00","value":365,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"SNAP-ChIP K-AcylStat Panel Spike-in Normalization Control for Chromatin IP (ChIP) and ChIP-Seq","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"shipping_messages":[],"rating":0,"meta_keywords":"SNAP-ChIP Barcoded Designer Nucleosomes for Chromatin IP Experiments, spike-in, chromatin IP, ChIP, chromatin immunoprecipitation, SNAP-ChIP, K-metstat, spike in, spike-in, drosophila spike-in, chip spike-in, chip spike in, chip normalization, chip-seq normalization, chip-seq spike-in, chip spike-in, chip-seq, quantitative chip spike-in, quantitative chip normalization, quantitative chip-seq normalization, quantitative chip spike-in, quantitative chip control, spike-in, drosophila spike-in, chromatin ip spike-in, chromatin ip spike in, chromatin ip normalization, chromatin ip-seq normalization, chromatin ip-seq spike-in, chromatin ip spike-in, chromatin ip-seq, quantitative chromatin ip spike-in, quantitative chromatin ip normalization, quantitative chromatin ip-seq normalization, quantitative chromatin ip spike-in, quantitative chromatin ip control, spike-in, drosophila spike-in, chromatin immunoprecipitation spike-in, chromatin immunoprecipitation spike in, chromatin immunoprecipitation normalization, chromatin immunoprecipitation-seq normalization, chromatin immunoprecipitation-seq spike-in, chromatin immunoprecipitation spike-in, chromatin immunoprecipitation-seq, quantitative chromatin immunoprecipitation spike-in, quantitative chromatin immunoprecipitation normalization, quantitative chromatin immunoprecipitation-seq normalization, quantitative chromatin immunoprecipitation spike-in, quantitative chromatin immunoprecipitation control, ChIP-seq calibration, chromatin IP calibration, ChIP calibration, chromatin immunoprecipitation calibration, ChIP-seq normalization, chromatin IP normalization, ChIP normalization, chromatin immunoprecipitation normalization, ChIP-seq spike-in, chromatin IP spike-in, ChIP spike-in, chromatin immunoprecipitation spike-in, ChIP-seq spike in, chromatin IP spike in, ChIP spike in, chromatin immunoprecipitation spike in, ChIP-seq antibody specificity, chromatin IP antibody specificity, ChIP antibody specificity, chromatin immunoprecipitation antibody specificity, histone acylation, crotonylation, butyrlation, acetylation, 19-3001, snap-ChIP K-AcylaStat, spike-in","show_quantity_input":1,"AddThisServiceButtonMeta":"","videos":{"list":[{"id":"-AlJ7Rv3beo","title_short":"An Introductio...","title_long":"An Introduction to SNAP-ChIP® Spike-in Controls","description_long":"UPDATED VIDEO: https://youtu.be/ElUS2NG5WyE.\nSNAP-ChIP (Sample Normalization and Antibody Profiling for Chromatin Immunoprecipitation) is a groundbreaking new technology that allows you to normalize your ChIP and ChIP-seq experiments as well as profile the specificity of your ChIP antibodies. Learn more about SNAP-ChIP® Spike-in Control Panels at www.epicypher.com.","description_short":"UPDATED VIDEO: https://youtu.be/ElUS2NG5WyE.\nSNAP-ChIP (Sample...","length":"03:00"}],"featured":{"id":"-AlJ7Rv3beo","title_short":"An Introductio...","title_long":"An Introduction to SNAP-ChIP® Spike-in Controls","description_long":"UPDATED VIDEO: https://youtu.be/ElUS2NG5WyE.\nSNAP-ChIP (Sample Normalization and Antibody Profiling for Chromatin Immunoprecipitation) is a groundbreaking new technology that allows you to normalize your ChIP and ChIP-seq experiments as well as profile the specificity of your ChIP antibodies. Learn more about SNAP-ChIP® Spike-in Control Panels at www.epicypher.com.","description_short":"UPDATED VIDEO: https://youtu.be/ElUS2NG5WyE.\nSNAP-ChIP (Sample...","length":"03:00"}},"title":"SNAP-ChIP® K-AcylStat Panel","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/687/654/SNAP_Chip__17660.1557847064.png?c=2","alt":"SNAP-ChIP® K-AcylStat Panel"}]}
{"sku":"19-3001","url":"https://www.epicypher.com/products/nucleosomes/snap-chip-k-acylstat-panel","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"options":[{"id":704,"type":"Configurable_PickList_Set","display_name":"Select Pack Size:","required":true,"condition":true,"state":"variant_option","values":[{"label":"20 µl","id":111,"data":"20 µl","selected":true},{"label":"200 µl","id":106,"data":"200 µl","selected":false}],"partial":"set-radio"}],"id":687,"bulk_discount_rates":[],"can_purchase":true,"meta_description":"SNAP-ChIP K-AcylStat Panel Spike-in Normalization Control for Chromatin IP (ChIP) and ChIP-Seq","category":["Nucleosomes","Nucleosomes/SNAP-ChIP<sup>®</sup> Spike-ins"],"main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/687/654/SNAP_Chip__17660.1557847064.png?c=2","alt":"SNAP-ChIP® K-AcylStat Panel"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=687","shipping":{"calculated":true},"num_reviews":0,"weight":"0.01 LBS","description":"<table border=\"0\" cellpadding=\"2\" cellspacing=\"2\" width=\"100%\">\n\t<tbody>\n\t\t<tr valign=\"top\">\n\t\t\t<td>\n\t\t\t<table border=\"0\" cellpadding=\"1\" cellspacing=\"1\" height=\"1490\" width=\"90%\">\n\t\t\t\t<tbody>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td valign=\"top\">\n\t\t\t\t\t\t<p></p>\n\n\t\t\t\t\t\t\n\t\t\t\t\t\t<span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b><b>SNAP-ChIP K-AcylStat Panel</b> Description:</b> A panel of distinctly modified mononucleosomes assembled from recombinant human histones expressed in E. coli (two each of histones H2A, H2B, H3 and H4; accession numbers: H2A-P04908; H2B-O60814; H3.1-P68431 or H3.2-Q71DI3*; H4-P62805) wrapped by 147 base pairs of barcoded Widom 601 positioning sequence DNA. The panel is comprised of a pool of 1 unmodified plus 22 histone H2A, H3, or H4 post-translational modifications (PTMs, created by a proprietary semi-synthetic method): H3K4ac, H3K9ac, H3K14ac, H3K18ac, H3K23ac, H3K27ac, H3K36ac, H4K5ac, H4K8ac, H4K12ac, H4K16ac, H4K20ac, H3K9bu, H3K9cr, H3K18bu, H3K18cr, H3K27bu, H3K27cr, H3K27acS28phos, H3K4,9,14,18ac, H4K5,8,12,16ac, H2AK5,8,13,15ac. Each distinctly modified nucleosome is distinguishable by a unique sequence of DNA (“barcode”) at the 3’ end that can be deciphered by qPCR or next-generation sequencing. Each of the 23 nucleosomes in the pool is wrapped by 2 distinct DNA species, each containing a distinct barcode (”A” and “B”, see SNAP-ChIP Manual) allowing for an internal technical replicate.</span></span>\n\n\t\t\t\t\t\t<p><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><span style=\"color: orange !important;\"><b>Learn more about SNAP-ChIP Certified Antibodies <a href=\"https://www.epicypher.com/snap-chip-certified-antibodies/\">here</a></b></span></span></span></p>\n\n\t\t\t\t\t\t<p><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\">* Histone H3.2 contains a Cys to Ala mutation at position 110. </span></span><br />\n\t\t\t\t\t\t</p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td valign=\"top\"><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b><b>SNAP-ChIP K-AcylStat Panel</b> Formulation:</b> PPurified recombinant mononucleosomes, containing a mixture of 23 (1 unmodified plus 22 unique) H2A, H3, and H4 PTMs in 10 mM sodium cacodylate, pH 7.5, 100 mM NaCl, 1 mM EDTA, 50% glycerol (w/v), 1x Protease Inhibitor cocktail, 100 µg/mL BSA, 10 mM β-mercaptoethanol. Average molarity = 0.6 nM. MW = ~199382.1 Da (average MW of all 23 nucleosomes).</span></span><br /><br />\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td valign=\"top\"><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b>SNAP-ChIP K-AcylStat Panel Storage and Stability:</b> Stable for six months at -20°C from date of receipt.</span></span><br /><br />\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td valign=\"top\"><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b>SNAP-ChIP K-AcylStat Panel Application Notes:</b> SNAP-ChIP K-AcylStat barcoded nucleosome standards are highly purified recombinant mononucleosomes and are suitable for use as spike-in controls for ChIP reactions, for antibody specificity testing or for effector protein binding experiments. See product manual for more information. SNAP-ChIP can easily be introduced into existing ChIP and ChIP-seq workflows for experiment normalization and monitoring of technical variability. </span></span><br /><br />\n\t\t\t\t\t\t<p><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><span style=\"color: orange !important;\"><b>Request a quote <a href=\"https://www.epicypher.com/request-a-quote/\">here</a></b></span></span></span></p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td valign=\"top\">\n\t\t\t\t\t\t<p><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b>Background References</b>:<br />\n\t\t\t\t\t\tGrzybowski A et al. 2015 Mol Cell 58: 886 - 899 <a href=\"https://www.ncbi.nlm.nih.gov/pubmed/26004229\" target=\"_new\" title=\"Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide\">LINK</a></span></span></p>\n\n\t\t\t\t\t\t<p><span style=\"font-size:12pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b>Product References</b>:<br />\n\t\t\t\t\t\tTay RE et al. 2020 J Exp. Med. 217: e20191453 <a href=\"https://www.ncbi.nlm.nih.gov/pubmed/32374402\" target=\"_new\" title=\"Hdac3 is an epigenetic inhibitor of the cytotoxicity program in CD8 T cells\">LINK</a></span></span><br />\n\t\t\t\t\t\t</p>\n\n\t\t\t\t\t\t<p>\n\t\t\t\t\t\t</p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td valign=\"top\">\n\t\t\t\t\t\t<p><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><b>SNAP-ChIP K-AcylStat Panel Additional Resources and Files:</b></span></span></p>\n\n\t\t\t\t\t\t<table border=\"0\" cellpadding=\"0\" cellspacing=\"10\" height=\"447\" width=\"450\">\n\t\t\t\t\t\t\t<tbody>\n\t\t\t\t\t\t\t\t<tr>\n\t\t\t\t\t\t\t\t\t<td style=\"height:60px\" valign=\"top\" width=\"350\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>Technical Datasheet </strong></span></span></td>\n\t\t\t\t\t\t\t\t\t<td align=\"center\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/documents/tds/19-3001.pdf\" target=\"_new\"><img alt=\"19-3001 Datasheet\" height=\"40\" src=\"/content/documents/tds/icon.png\" width=\"30\" /></a></span></span></td>\n\t\t\t\t\t\t\t\t</tr>\n\t\t\t\t\t\t\t\t<tr>\n\t\t\t\t\t\t\t\t\t<td style=\"height:60px\" valign=\"top\" width=\"350\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>SNAP-ChIP K-AcylStat Manual</strong></span></span></td>\n\t\t\t\t\t\t\t\t\t<td align=\"center\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/documents/snapchip/SNAP-ChIP_K-AcylStat_Manual.pdf\" target=\"_new\"><img alt=\"SNAP-ChIP K-AcylStat manual.pdf\" height=\"40\" src=\"/content/documents/tds/icon.png\" width=\"30\" /></a></span></span></td>\n\t\t\t\t\t\t\t\t</tr>\n\t\t\t\t\t\t\t\t<tr>\n\t\t\t\t\t\t\t\t\t<td style=\"height:60px\" valign=\"top\" width=\"350\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>SNAP-ChIP Brochure</strong></span></span></td>\n\t\t\t\t\t\t\t\t\t<td align=\"center\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/docs/SNAP-ChIP.pdf\" target=\"_new\"><img alt=\"SNAP-ChIP K-AcylStat manual.pdf\" height=\"40\" src=\"/content/documents/tds/icon.png\" width=\"30\" /></a></span></span></td>\n\t\t\t\t\t\t\t\t</tr>\n\t\t\t\t\t\t\t\t<tr>\n\t\t\t\t\t\t\t\t\t<td style=\"height:60px\" valign=\"top\" width=\"350\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>SNAP-ChIP K-AcylStat Full Panel Information: </strong>Forward/reverse barcodes, nucleosomal DNA sequences, qPCR primer sequences, TaqMan probe sequence</span></span></td>\n\t\t\t\t\t\t\t\t\t<td align=\"center\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/documents/snapchip/SNAP-ChIP_K-AcylStat_19-3001_Barcode-Sequence-Primer_Info-SHORT_ver3.0.xlsx\" target=\"_new\"><img alt=\"19-3001 Datasheet\" height=\"40\" src=\"/content/documents/excelicon.png\" width=\"30\" /></a></span></span></td>\n\t\t\t\t\t\t\t\t</tr>\n\t\t\t\t\t\t\t\t<tr>\n\t\t\t\t\t\t\t\t\t<td style=\"height:60px\" valign=\"top\" width=\"350\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>SNAP-ChIP K-AcylStat qPCR Primer Sequences</strong> - Formatted for easy upload for ordering from common vendors (<em>e.g.</em> IDT)</span></span></td>\n\t\t\t\t\t\t\t\t\t<td align=\"center\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/documents/snapchip/SNAP-ChIP_K-AcylStat_qPCR_Primers_BarcodeA_v2.0.xlsx\" target=\"_new\"><img alt=\"19-3001 Datasheet\" height=\"40\" src=\"/content/documents/excelicon.png\" width=\"30\" /></a></span></span></td>\n\t\t\t\t\t\t\t\t</tr>\n\t\t\t\t\t\t\t\t<tr>\n\t\t\t\t\t\t\t\t\t<td style=\"height:60px\" valign=\"top\" width=\"350\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>Fasta formatted file of K-AcylStat barcode Sequences</strong> - For alignment of NGS sequencing data</span></span></td>\n\t\t\t\t\t\t\t\t\t<td align=\"center\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/documents/snapchip/KacylStat_barcoded_fasta_v2.0.rtf\" target=\"_new\"><img alt=\"19-3001 fasta\" height=\"40\" src=\"/content/documents/excelicon.png\" width=\"30\" /></a></span></span></td>\n\t\t\t\t\t\t\t\t</tr>\n\t\t\t\t\t\t\t</tbody>\n\t\t\t\t\t\t</table>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t</tbody>\n\t\t\t</table>\n\t\t\t</td>\n\t\t\t<td>\n\t\t\t<table border=\"0\" cellpadding=\"3\" cellspacing=\"3\" width=\"100%\">\n\t\t\t\t<tbody>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"center\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/images/nucleosome/k-acylstat_panel.gif\" target=\"_new\"><img alt=\"K-AcylStat Panel\" src=\"/content/images/nucleosome/k-acylstat_panel.gif\" width=\"250\" /></a></span></span></td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"left\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>SNAP-ChIP K-AcylStat Panel:</strong> Schematic representation of nucleosomes comprising the K-AcylStat Panel: H3K4ac, H3K9ac, H3K14ac, H3K18ac, H3K23ac, H3K27ac, H3K36ac, H4K5ac, H4K8ac, H4K12ac, H4K16ac, H4K20ac, H3K9bu, H3K9cr, H3K18bu, H3K18cr, H3K27bu, H3K27cr, H3K27acS28phos, H3K4,9,14,18ac, H4K5,8,12,16ac and unmodified.<br />\n\t\t\t\t\t\t<strong>(Click image to enlarge) </strong></span></span>\n\t\t\t\t\t\t<p></p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t</tbody>\n\t\t\t</table>\n\n\t\t\t<table border=\"0\" cellpadding=\"3\" cellspacing=\"3\" width=\"100%\">\n\t\t\t\t<tbody>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"center\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/images/products/nucleosomes/19-3001-DNA-gel.jpg\" target=\"_new\"><img alt=\"19-3001 DNA Gel Data\" src=\"/content/images/products/nucleosomes/19-3001-DNA-gel.jpg\" width=\"250\" /></a></span></span></td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"left\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>DNA Gel Data:</strong> Representative images for SNAP-ChIP K-AcylStats resolved by native PAGE and stained with ethidium bromide to visualize DNA. Lane 1: Free 147bp DNA used in nuclesome assembly (100 ng). Lane 2: Intact nucleosomes (200 ng). Comparable experiments were performed for the entire K-AcylStat Panel.<br />\n\t\t\t\t\t\t<strong>(Click image to enlarge) </strong></span></span>\n\t\t\t\t\t\t<p></p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t</tbody>\n\t\t\t</table>\n\n\t\t\t<table border=\"0\" cellpadding=\"3\" cellspacing=\"3\" width=\"100%\">\n\t\t\t\t<tbody>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"center\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/images/products/nucleosomes/19-3001-protein-gel.jpg\" target=\"_new\"><img alt=\"19-3001 Protein Gel Data\" src=\"/content/images/products/nucleosomes/19-3001-protein-gel.jpg\" width=\"250\" /></a></span></span></td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"left\" valign=\"top\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>Protein Gel Data:</strong> Representative Coomassie stained PAGE gel of SNAP-ChIP K-AcylStat (2 µg each) to demonstrate the purity of the histones in the preparation. Sizes of molecular weight markers and positions of the core histones (H2A, H2B, H3 and H4) are indicated. Comparable experiments were performed for the entire K-AcylStat Panel.<br />\n\t\t\t\t\t\t<strong>(Click image to enlarge) </strong></span></span>\n\t\t\t\t\t\t<p></p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t</tbody>\n\t\t\t</table>\n\n\t\t\t<table border=\"0\" cellpadding=\"3\" cellspacing=\"3\" width=\"100%\">\n\t\t\t\t<tbody>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"center\"><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><a href=\"/content/images/products/nucleosomes/19-3001-ChIP-data.jpg\" target=\"_new\"><img alt=\"19-3001 ChIP Data\" src=\"/content/images/products/nucleosomes/19-3001-ChIP-data.jpg\" width=\"250\" /></a></span></span></td>\n\t\t\t\t\t</tr>\n\t\t\t\t\t<tr>\n\t\t\t\t\t\t<td align=\"left\" valign=\"top\">\n\t\t\t\t\t\t<p><span style=\"font-size:11pt;\"><span style=\"font-family:arial,helvetica,sans-serif;\"><strong>ChIP Data:</strong> Representative chromatin imunoprecipitation (ChIP) data using commerically available ChIP-grade antibodies targeting the indicated PTMs. The antibodies were assayed in a native ChIP experiment with 3 μg antibody added to 3 μg K-562 chromatin with the K-AcylStat Panel spiked-in prior to micrococcal nuclease digestion. Quantitative real-time PCR (qPCR) was used to measure recovery of duplicate DNA barcodes corresponding to each uniquely modified nucleosome in the panel (blue bars, X-axis). The black bars map to the log scale on the right y-axis and indicate the percentage of target immunoprecipitated relative to the input (a measure of the antibody efficiency). In each case, the SNAP-ChIP spike-in confirmed that the antibodies recovered the expected histone PTM with high efficiency and specificity. Comparable experiments were performed for the entire K-AcylStat Panel.<br />\n\t\t\t\t\t\t<strong>(Click image to enlarge) </strong></span></span></p>\n\n\t\t\t\t\t\t<p></p>\n\t\t\t\t\t\t</td>\n\t\t\t\t\t</tr>\n\t\t\t\t</tbody>\n\t\t\t</table>\n\n\t\t\t<table border=\"0\" cellpadding=\"3\" cellspacing=\"3\" width=\"100%\">\n\t\t\t\t<tbody>\n\t\t\t\t</tbody>\n\t\t\t</table>\n\t\t\t</td>\n\t\t</tr>\n\t</tbody>\n</table>\n\n","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$365.00","value":365,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"SNAP-ChIP K-AcylStat Panel Spike-in Normalization Control for Chromatin IP (ChIP) and ChIP-Seq","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"shipping_messages":[],"rating":0,"meta_keywords":"SNAP-ChIP Barcoded Designer Nucleosomes for Chromatin IP Experiments, spike-in, chromatin IP, ChIP, chromatin immunoprecipitation, SNAP-ChIP, K-metstat, spike in, spike-in, drosophila spike-in, chip spike-in, chip spike in, chip normalization, chip-seq normalization, chip-seq spike-in, chip spike-in, chip-seq, quantitative chip spike-in, quantitative chip normalization, quantitative chip-seq normalization, quantitative chip spike-in, quantitative chip control, spike-in, drosophila spike-in, chromatin ip spike-in, chromatin ip spike in, chromatin ip normalization, chromatin ip-seq normalization, chromatin ip-seq spike-in, chromatin ip spike-in, chromatin ip-seq, quantitative chromatin ip spike-in, quantitative chromatin ip normalization, quantitative chromatin ip-seq normalization, quantitative chromatin ip spike-in, quantitative chromatin ip control, spike-in, drosophila spike-in, chromatin immunoprecipitation spike-in, chromatin immunoprecipitation spike in, chromatin immunoprecipitation normalization, chromatin immunoprecipitation-seq normalization, chromatin immunoprecipitation-seq spike-in, chromatin immunoprecipitation spike-in, chromatin immunoprecipitation-seq, quantitative chromatin immunoprecipitation spike-in, quantitative chromatin immunoprecipitation normalization, quantitative chromatin immunoprecipitation-seq normalization, quantitative chromatin immunoprecipitation spike-in, quantitative chromatin immunoprecipitation control, ChIP-seq calibration, chromatin IP calibration, ChIP calibration, chromatin immunoprecipitation calibration, ChIP-seq normalization, chromatin IP normalization, ChIP normalization, chromatin immunoprecipitation normalization, ChIP-seq spike-in, chromatin IP spike-in, ChIP spike-in, chromatin immunoprecipitation spike-in, ChIP-seq spike in, chromatin IP spike in, ChIP spike in, chromatin immunoprecipitation spike in, ChIP-seq antibody specificity, chromatin IP antibody specificity, ChIP antibody specificity, chromatin immunoprecipitation antibody specificity, histone acylation, crotonylation, butyrlation, acetylation, 19-3001, snap-ChIP K-AcylaStat, spike-in","show_quantity_input":1,"AddThisServiceButtonMeta":"","videos":{"list":[{"id":"-AlJ7Rv3beo","title_short":"An Introductio...","title_long":"An Introduction to SNAP-ChIP® Spike-in Controls","description_long":"UPDATED VIDEO: https://youtu.be/ElUS2NG5WyE.\nSNAP-ChIP (Sample Normalization and Antibody Profiling for Chromatin Immunoprecipitation) is a groundbreaking new technology that allows you to normalize your ChIP and ChIP-seq experiments as well as profile the specificity of your ChIP antibodies. Learn more about SNAP-ChIP® Spike-in Control Panels at www.epicypher.com.","description_short":"UPDATED VIDEO: https://youtu.be/ElUS2NG5WyE.\nSNAP-ChIP (Sample...","length":"03:00"}],"featured":{"id":"-AlJ7Rv3beo","title_short":"An Introductio...","title_long":"An Introduction to SNAP-ChIP® Spike-in Controls","description_long":"UPDATED VIDEO: https://youtu.be/ElUS2NG5WyE.\nSNAP-ChIP (Sample Normalization and Antibody Profiling for Chromatin Immunoprecipitation) is a groundbreaking new technology that allows you to normalize your ChIP and ChIP-seq experiments as well as profile the specificity of your ChIP antibodies. Learn more about SNAP-ChIP® Spike-in Control Panels at www.epicypher.com.","description_short":"UPDATED VIDEO: https://youtu.be/ElUS2NG5WyE.\nSNAP-ChIP (Sample...","length":"03:00"}},"title":"SNAP-ChIP® K-AcylStat Panel","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/687/654/SNAP_Chip__17660.1557847064.png?c=2","alt":"SNAP-ChIP® K-AcylStat Panel"}]}
|
|
Current stock:
0