H3.3S31phos,K36me3 Recombinant Nucleosome, Biotinylated
{"url":"https://www.epicypher.com/products/nucleosomes/modified-designer-nucleosomes-dnucs/h3-3s31phos-k36me3-recombinant-nucleosome-biotinylated","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"options":[],"id":1153,"bulk_discount_rates":[],"can_purchase":false,"meta_description":"Designer nucleosome (dNuc) H3.3S31phosK38me3 is highly purified and suitable for a variety of applications, including use as a substrate in enzyme assays, high-throughput screening and inhibitor testing, chromatin binding studies, protein-protein interaction assays, structural studies, and in effector protein binding experiments.","category":["Nucleosomes","Nucleosomes/Modified Designer Nucleosomes (dNucs™)"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1153/1226/Recombinant_nucleosome__69501.1717617797.png?c=2","alt":"H3.3S31phos,K36me3 Recombinant Nucleosome, Biotinylated"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1153","shipping":{"calculated":true},"num_reviews":0,"weight":"0.01 LBS","custom_fields":[{"id":"1319","name":"Pack Size","value":"50 μg"}],"sku":"16-0407","description":"<div class=\"product-general-info\">\n <ul class=\"product-general-info__list-left\">\n <li class=\"product-general-info__list-item\">\n <strong>Species: </strong>Human\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Source: </strong><i>E. coli</i> & synthetic DNA\n </li>\n </ul>\n <ul class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <strong>Tag: </strong>Biotinylated\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Molecular Weight: </strong>199,836 Da\n </li>\n </ul>\n</div>\n<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <p>\n Histone phosphorylation is a post-translational modification (PTM) wherein a phosphate group is added to a\n histone protein, predominantly occurring on serine, threonine, and tyrosine residues. In combination with\n other PTMs, histone phosphorylation constitutes the “histone code,” acting as a language read by proteins to\n regulate chromatin structure and gene expression. Histone phosphorylation is involved in chromatin\n remodeling and compaction associated with diverse cellular processes, including DNA damage repair,\n transcription regulation, cell division, and apoptosis [1]. Histone phosphorylation is also observed on\n non-canonical histones, particularly H3.3, where it plays roles in transcriptional regulation. Recombinant\n mononucleosomes containing phosphorylated histones can be used to study the biological functions of histone\n phosphorylation.\n </p>\n <p>\n H3.3S31phos,K36me3 (histone H3.3 serine 31 phosphorylation, lysine 36 trimethylation) Recombinant\n Nucleosome, Biotinylated consists of 147 base pairs of DNA wrapped around an octamer of core histone\n proteins (two each of H2A, H2B, H3.3, and H4) to form a nucleosome, the basic repeating unit of chromatin.\n The 147 bp 601 sequence, identified by Lowary and Widom [2], has high affinity for histone octamers and is\n useful for nucleosome assembly. The DNA contains a 5’ biotin-TEG group. The variant H3.3 contains a serine\n at position 31, one of several discrete amino acid differences compared to canonical H3.1.\n H3.3S31phos,K36me3 contains phosphorylated serine at position 31 and trimethylated lysine at position 36 on\n histone H3.3. H3.3S31phos,K36me3 modulates stimulation-responsive gene expression in activated macrophages\n via engagement of the histone methyltransferase SETD2 and ejection of the transcriptional co-repressor\n ZMYND11 [3].\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/nucleosomes/16-0407-protein-gel-data.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\">\n <img alt=\"16-0407-protein-gel-data\"\n src=\"/content/images/products/nucleosomes/16-0407-protein-gel-data.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span>\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 1: Protein gel data </strong><br />\n Coomassie stained PAGE gel of proteins in H3.3S31phos,K36me3 nucleosome (1 µg) demonstrates the\n purity of histones in the preparation. Sizes of molecular weight markers and positions of the core\n histones (H2A, H2B, H3.3S31phos,K36me3, and H4) are indicated.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/nucleosomes/16-0407-mass-spec-data.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img alt=\"16-0407-mass-spec-data\"\n src=\"/content/images/products/nucleosomes/16-0407-mass-spec-data.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 2: Mass spec data </strong><br />\n Synthetic H3.3S31phos,K36me3 histone analyzed by high resolution mass spectrometry. Expected mass =\n 15,319.7 Da. Determined mass = 15,316.86 Da.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/nucleosomes/16-0407-dna-gel-data.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img alt=\"16-0407-dna-gel-data\"\n src=\"/content/images/products/nucleosomes/16-0407-dna-gel-data.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 3: DNA gel data </strong><br />\n H3.3S31phos,K36me3 nucleosome resolved via native PAGE and stained with\n ethidium bromide to visualize DNA. Both lanes are from the same gel.\n <strong>Lane 1:</strong> Free DNA (EpiCypher\n <a href=\"/products/nucleosomes/nucleosome-assembly-601-sequence-dna-biotinylated\"\n target=\"_blank\">18-0005</a>; 100 ng). Free DNA is over 95% pure by densitometry. <strong>Lane 2:</strong> Intact H3.3S31phos,K36me3\n nucleosomes (400 ng).\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img alt=\"16-0407-protein-gel-data\"\n src=\"/content/images/products/nucleosomes/16-0407-protein-gel-data.jpeg\"\n class=\"image-picker__side-image\" role=\"button\" />\n <img alt=\"16-0407-mass-spec-data\" src=\"/content/images/products/nucleosomes/16-0407-mass-spec-data.jpeg\"\n class=\"image-picker__side-image\" role=\"button\" />\n <img alt=\"16-0407-dna-gel-data\" src=\"/content/images/products/nucleosomes/16-0407-dna-gel-data.jpeg\"\n class=\"image-picker__side-image\" role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Stable for six months at -80°C from date of receipt. For best\n results, aliquot and avoid freeze/thaws.\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Formulation</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 10 mM Tris-HCl pH 7.5, 25 mM NaCl, 1 mM EDTA, 2 mM DTT, 20%\n glycerol (27.2 µg protein, 50 µg DNA + protein).\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Application Notes</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <p>\n H3.3S31phos,K36me3 mononucleosome is highly purified and suitable for a variety of applications, including\n use as a substrate in enzyme assays, high-throughput screening and inhibitor testing, chromatin binding\n studies, protein-protein interaction assays, structural studies, and in effector protein binding\n experiments. For a corresponding unmodified control, we recommend EpiCypher <a\n href=\"/products/nucleosomes/mononucleosomes-recombinant-human-biotinylated\" target=\"_blank\">16-0006</a>.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Gene & Protein Information</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>UniProt ID</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n H2A - P04908 (alt. names: H2A type 1-B/E, H2A.2, H2A/a,\n H2A/m)<br />\n H2B - O60814 (alt. names: H2B K, HIRA-interacting protein 1)<br />\n H3.3 - P84243<br />\n H4 - P62805\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">References</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <strong>Background References:</strong>\n <br />\n [1] Rossetto et al. <em>Epigenetics</em> (2012). PMID:\n <a href=\"https://pubmed.ncbi.nlm.nih.gov/22948226/\" target=\"_blank\"\n title=\"Histone phosphorylation: a chromatin modification involved in diverse nuclear events\">22948226</a><br />\n [2] Lowary and Widom <em>J. Mol. Biol.</em> (1998). PMID:\n <a href=\"https://pubmed.ncbi.nlm.nih.gov/9514715/\" target=\"_blank\"\n title=\"New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning\">9514715</a><br />\n [3] Armache et al. <em>Nature</em> (2020). PMID:\n <a href=\"https://pubmed.ncbi.nlm.nih.gov/32699416/\" target=\"_blank\"\n title=\"Histone H3.3 phosphorylation amplifies stimulation-induced transcription\">32699416</a><br />\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Documents & Resources</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-documents\">\n <a href=\"/content/documents/tds/16-0407.pdf\" target=\"_blank\" class=\"product-documents__link\">\n <svg version=\"1.1\" id=\"Layer_1\" xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\" x=\"0px\" y=\"0px\" viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\" xml:space=\"preserve\" class=\"product-documents__icon\"\n alt=\"16-0407 Datasheet\">\n <g>\n <path class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect x=\"20\" y=\"112\" class=\"product-documents__svg-background\" width=\"146\" height=\"76\" />\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path class=\"product-documents__svg-pdf\" d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Technical Datasheet</span>\n </a>\n </div>\n </div>\n </div>\n </div>\n </div>\n</div>","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$575.00","value":575,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"H3.3S31phos,K36me3 Recombinant Nucleosome, Biotinylated","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"shipping_messages":[],"rating":0,"meta_keywords":"designer nucleosome, dNuc, H3.3S31phosK36me3, nucleosome, mononucleosome","show_quantity_input":1,"title":"H3.3S31phos,K36me3 Recombinant Nucleosome, Biotinylated","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1153/1226/Recombinant_nucleosome__69501.1717617797.png?c=2","alt":"H3.3S31phos,K36me3 Recombinant Nucleosome, Biotinylated"}]} Pack Size: 50 μg
- Species: Human
- Source: E. coli & synthetic DNA
- Tag: Biotinylated
- Molecular Weight: 199,836 Da
Description
Histone phosphorylation is a post-translational modification (PTM) wherein a phosphate group is added to a histone protein, predominantly occurring on serine, threonine, and tyrosine residues. In combination with other PTMs, histone phosphorylation constitutes the “histone code,” acting as a language read by proteins to regulate chromatin structure and gene expression. Histone phosphorylation is involved in chromatin remodeling and compaction associated with diverse cellular processes, including DNA damage repair, transcription regulation, cell division, and apoptosis [1]. Histone phosphorylation is also observed on non-canonical histones, particularly H3.3, where it plays roles in transcriptional regulation. Recombinant mononucleosomes containing phosphorylated histones can be used to study the biological functions of histone phosphorylation.
H3.3S31phos,K36me3 (histone H3.3 serine 31 phosphorylation, lysine 36 trimethylation) Recombinant Nucleosome, Biotinylated consists of 147 base pairs of DNA wrapped around an octamer of core histone proteins (two each of H2A, H2B, H3.3, and H4) to form a nucleosome, the basic repeating unit of chromatin. The 147 bp 601 sequence, identified by Lowary and Widom [2], has high affinity for histone octamers and is useful for nucleosome assembly. The DNA contains a 5’ biotin-TEG group. The variant H3.3 contains a serine at position 31, one of several discrete amino acid differences compared to canonical H3.1. H3.3S31phos,K36me3 contains phosphorylated serine at position 31 and trimethylated lysine at position 36 on histone H3.3. H3.3S31phos,K36me3 modulates stimulation-responsive gene expression in activated macrophages via engagement of the histone methyltransferase SETD2 and ejection of the transcriptional co-repressor ZMYND11 [3].
Validation Data
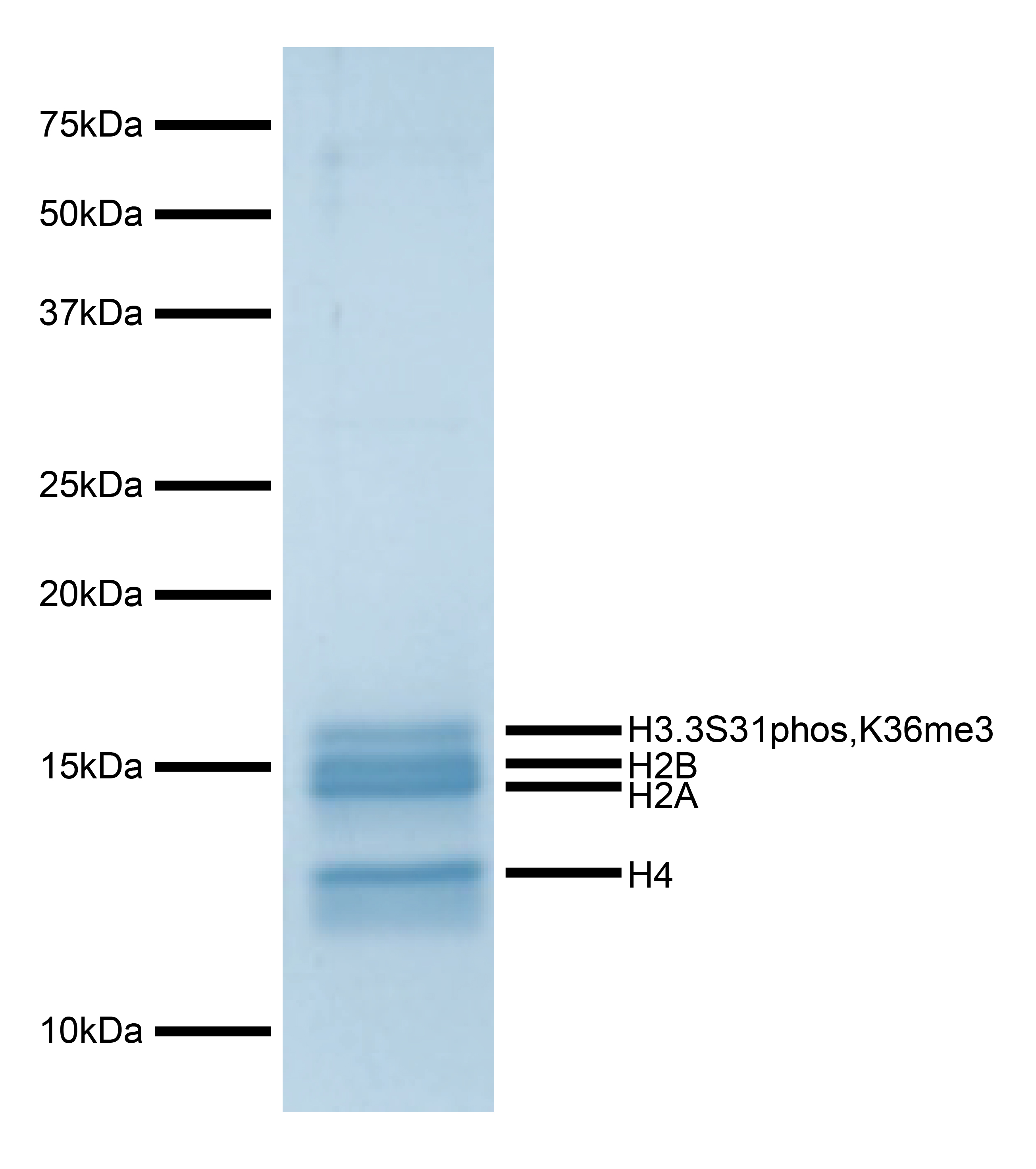
Figure 1: Protein gel data
Coomassie stained PAGE gel of proteins in H3.3S31phos,K36me3 nucleosome (1 µg) demonstrates the
purity of histones in the preparation. Sizes of molecular weight markers and positions of the core
histones (H2A, H2B, H3.3S31phos,K36me3, and H4) are indicated.
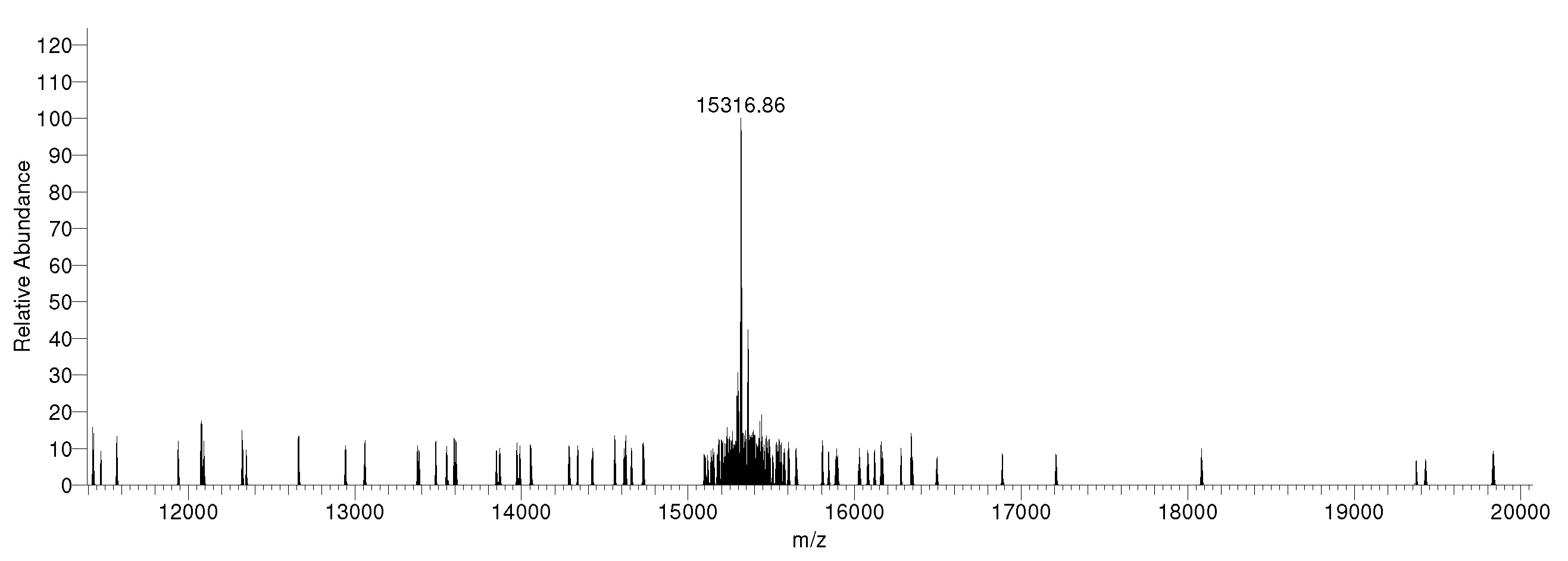
Figure 2: Mass spec data
Synthetic H3.3S31phos,K36me3 histone analyzed by high resolution mass spectrometry. Expected mass =
15,319.7 Da. Determined mass = 15,316.86 Da.
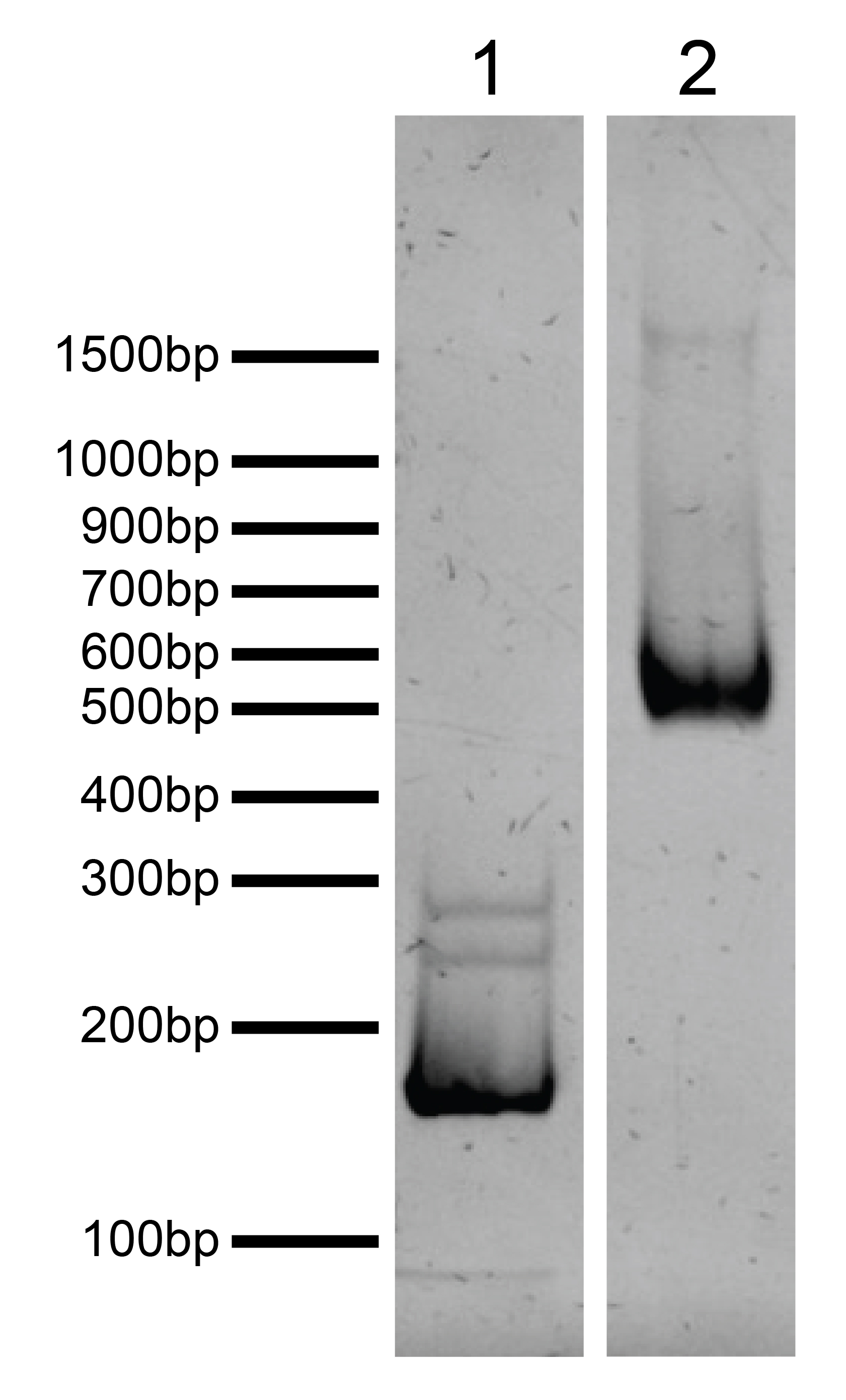
Figure 3: DNA gel data
H3.3S31phos,K36me3 nucleosome resolved via native PAGE and stained with
ethidium bromide to visualize DNA. Both lanes are from the same gel.
Lane 1: Free DNA (EpiCypher
18-0005; 100 ng). Free DNA is over 95% pure by densitometry. Lane 2: Intact H3.3S31phos,K36me3
nucleosomes (400 ng).
Technical Information
Application Notes
H3.3S31phos,K36me3 mononucleosome is highly purified and suitable for a variety of applications, including use as a substrate in enzyme assays, high-throughput screening and inhibitor testing, chromatin binding studies, protein-protein interaction assays, structural studies, and in effector protein binding experiments. For a corresponding unmodified control, we recommend EpiCypher 16-0006.
Gene & Protein Information
H2B - O60814 (alt. names: H2B K, HIRA-interacting protein 1)
H3.3 - P84243
H4 - P62805