dCypher™ Nucleosome Full Panel
{"sku":"16-9001","url":"https://www.epicypher.com/products/nucleosomes/dcypher-nucleosome-panels/dcypher-nucleosome-full-panel","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"options":[],"id":662,"bulk_discount_rates":[],"can_purchase":false,"meta_description":"dCypher Nucleosome Full Panel for Enzyme Screening and HTS Assays","category":["Nucleosomes/dCypher™ Nucleosome Panels"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/662/975/dcypher-nucleosome-full-panel__10296.1645734567.jpg?c=2","alt":"dCypher Nucleosome Full Panel"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=662","shipping":{"calculated":true},"num_reviews":0,"weight":"0.00 LBS","description":"<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <p>\n Understanding epigenetic reader proteins and their binding\n preference for post-translational modifications (PTMs) is key to\n unveiling how these proteins regulate genome processes and how their\n dysregulation may be contributing to disease. The dCypher™\n Nucleosome Panel enables access to epigenetic diversity in a\n physiologically relevant context, the nucleosome, where chromatin\n reader activity can be accurately interrogated. The 96-well plate\n contains recombinant mononucleosomes as well as appropriate\n controls. Human histones expressed in <em>E. coli</em> bearing\n single and combinatorial histone post-translational modifications\n (PTMs) are wrapped by 147 or 199 base pair DNA with a 5’ biotin-TEG\n group and a central 601-positioning sequence, identified by Lowary\n and Widom [1].\n </p>\n <p>\n All nucleosomes in the panel are subjected to EpiCypher’s rigorous\n quality control metrics, including: ESI-TOF mass spectrometry\n analysis of the modified histones, SDS-PAGE to confirm octamer\n composition and purity, native PAGE to confirm nucleosome assembly,\n and western blot analysis of the PTM, histone mutation, or histone\n variant (if applicable). For the full list of nucleosomes in the\n panel, including individual catalog numbers of full-size (50 µg)\n products, see the Documents & Resources section for the associated\n Excel sheet.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div\n class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/nucleosomes/16-9001-dcypher-nuc-full-panel.jpeg\"\n target=\"_new\"\n class=\"image-picker__main-image-link\"\n ><img\n alt=\"16-9001-dcypher-nuc-full-panel\"\n src=\"/content/images/products/nucleosomes/16-9001-dcypher-nuc-full-panel.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong\n >Figure 1: dCypher nucleosome full panel plate layout and\n key </strong\n ><br />\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/nucleosomes/16-9001-binding-data.jpeg\"\n target=\"_new\"\n class=\"image-picker__main-image-link\"\n ><img\n alt=\"16-9001-binding-data\"\n src=\"/content/images/products/nucleosomes/16-9001-binding-data.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong>Figure 2: Chromatin reader binding data</strong\n ><br />\n GST-tagged HP1β (100 nM, EpiCypher\n <a\n href=\"https://www.epicypher.com/products/recombinant-proteins/hp1-recombinant-human-his-tagged\"\n >15-0074</a\n >) was assayed with AlphaScreen (PerkinElmer) technology to\n measure binding to nucleosomes (x-axis) in the dCypher\n nucleosome Panel (10 nM). HP1β shows binding to the H3K9me3\n and H3K9me2 dNucs, consistent with its reported binding\n preference [5].\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img\n alt=\"16-9001-dcypher-nuc-full-panel\"\n src=\"/content/images/products/nucleosomes/16-9001-dcypher-nuc-full-panel.jpeg\"\n width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\" />\n <img\n alt=\"16-9001-binding-data\"\n src=\"/content/images/products/nucleosomes/16-9001-binding-data.jpeg\"\n class=\"image-picker__side-image\"\n role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Stable for six months at -80°C from date of receipt. Avoid\n freeze/thaws.\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Formulation</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n The material included in this 96-well plate is provided at 1.5\n µM in 5 µL of nucleosome storage buffer (10 mM Tris-HCl pH 7.5,\n 25 mM NaCl, 1 mM EDTA, 2 mM DTT*, & 20% glycerol). *dNucs\n containing crotonyl-lysine are stored in nucleosome storage\n buffer without DTT. MW = ~200,000 Da\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Gene & Protein Information</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Uniprot ID</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n H2A - P04908 (alt. names: H2A type 1-B/E, H2A.2, H2A/a, H2A/m)\n <br />\n H2B - O60814 (alt. names: H2B K, HIRA-interacting protein 1)\n <br />\n H3.1 - P68431 (alt. names: H3, H3/a, H3/b, H3/c, H3/d)\n <br />\n H3.2 - Q71DI3\n <br />\n H3.3 - P84243\n <br />\n H4 - P62805\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Application Notes</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <p>\n Access to epigenetic diversity in the context of a physiological\n nucleosome enables broad end-user applications, including nucleosome\n binding studies (e.g. chromatin reader binding preferences [2-4] see\n <strong>Figure 2</strong>), enzyme screening assays (e.g.\n identification of preferred substrates), and antibody specificity\n testing (e.g. for applications where histone peptide specificity is\n an insufficient surrogate). The biotin group on the DNA facilitates\n a wide variety of applications involving streptavidin capture.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Frequently Asked Questions</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <p>\n <strong>Q:</strong>\n <em>How many assay points of each nucleosome are supplied?</em>\n <br />\n <strong>A:</strong> This varies according to the sensitivity of the\n end application. For some context, the supplied format is sufficient\n to perform >35 AlphaScreen assays, >30 IP-qPCR experiments, and 3-5\n immunoblotting experiments.\n </p>\n <p>\n <strong>Q:</strong>\n <em>How do I store the plate? Can I dilute the nucleosomes?</em>\n <br />\n <strong>A:</strong> The nucleosomes are stable for 6 months at -80ºC\n in the supplied format (1.5 µM). Freeze/thaw cycles should be\n limited (2-3 times). When diluted below 1.5 µM, a carrier protein\n (e.g. BSA) must be used to supplement the concentration, otherwise,\n stability is affected. For example, in our SNAP-ChIP product line,\n nucleosomes are diluted to 0.6 nM in 10 mM sodium cacodylate pH 7.5,\n 100 mM NaCl, 1 mM EDTA, 50% glycerol (w/v), 1x Protease Inhibitor\n cocktail, 100 μg/mL BSA, 10 mM β-mercaptoethanol. In this\n formulation, the nucleosomes are stable at -20ºC (where they do not\n freeze) for 6 months.\n </p>\n <p>\n For applications that may not be compatible with the SNAP-ChIP\n buffer, such as AlphaScreen, EpiCypher routinely dilutes nucleosomes\n to 80 nM in assay buffer (20 mM Tris pH 7.5, 0.01% BSA, 0.01% NP-40,\n 2mM DTT). Under these conditions the nucleosomes are stable for ~2\n weeks at -80°C.\n </p>\n <p>\n <strong>*Note:</strong> extremely high amounts of salt (>650 mM) and\n ionic detergents (e.g. SDS) will disrupt nucleosome stability.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">References</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <b>Background References:</b><br />\n [1] Lowary & Widom <em>J. Mol. Biol.</em> (1998). PMID:\n <a\n href=\"https://pubmed.ncbi.nlm.nih.gov/9514715/\"\n title=\"New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning\"\n target=\"_blank\"\n >9514715</a\n ><br />\n [2] Weinberg et al. <em>Nature</em> (2019). PMID:\n <a\n href=\"https://pubmed.ncbi.nlm.nih.gov/31485078/\"\n title=\"The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape\"\n target=\"_blank\"\n >31485078</a\n ><br />\n [3] Weinberg et al. <em>Nat. Genet.</em> (2021). PMID:\n <a\n href=\"https://pubmed.ncbi.nlm.nih.gov/33986537/\"\n title=\"Two competing mechanisms of DNMT3A recruitment regulate the dynamics of de novo DNA methylation at PRC1-targeted CpG islands\"\n target=\"_blank\"\n >33986537</a\n ><br />\n [4] Dilworth et al. <em>Nat. Chem. Biol</em> (2022). PMID:\n <a\n title=\"A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization\"\n href=\"https://pubmed.ncbi.nlm.nih.gov/34782742/\"\n target=\"_blank\"\n >34782742</a\n >\n <br />\n [5] Albanese et al. <em>ACS Chem. Biol.</em> (2020). PMID:\n <a\n title=\"Engineered Reader Proteins for Enhanced Detection of Methylated Lysine on Histones\"\n href=\"https://pubmed.ncbi.nlm.nih.gov/31634430/\"\n target=\"_blank\"\n >31634430</a\n >\n <br />\n <br />\n <strong>Product References:</strong>\n <iframe\n class=\"airtable-embed\"\n src=\"https://airtable.com/embed/shrFyHyvR9bSFrIBP?backgroundColor=purple&viewControls=on\"\n frameborder=\"0\"\n onmousewheel=\"\"\n width=\"100%\"\n height=\"300\"\n style=\"background: transparent; border: 1px solid #ccc\"></iframe>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Documents & Resources</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-documents\">\n <a\n href=\"/content/documents/tds/16-9001.pdf\"\n target=\"_new\"\n class=\"product-documents__link\">\n <svg\n version=\"1.1\"\n id=\"Layer_1\"\n xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\"\n x=\"0px\"\n y=\"0px\"\n viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\"\n xml:space=\"preserve\"\n class=\"product-documents__icon\"\n alt=\"16-0002 Datasheet\">\n <g>\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect\n x=\"20\"\n y=\"112\"\n class=\"product-documents__svg-background\"\n width=\"146\"\n height=\"76\" />\n <g>\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Technical Datasheet</span>\n </a>\n </div>\n <div class=\"product-documents\">\n <a\n href=\"/content/documents/16-9001-dcypher-nuc-panel-full-list.xlsx\"\n target=\"_new\"\n class=\"product-documents__link\">\n <svg\n version=\"1.1\"\n id=\"Layer_1\"\n xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\"\n x=\"0px\"\n y=\"0px\"\n viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\"\n xml:space=\"preserve\"\n class=\"product-documents__icon\">\n <g>\n <path\n class=\"product-documents__svg-excel\"\n d=\"M193.17,69.88l-46.83-46.83c-1.31-1.31-3.07-2.05-4.92-2.05H48.96C45.11,21,42,24.11,42,27.96v181.07\n c0,3.85,3.11,6.96,6.96,6.96h139.29c3.85,0,6.96-3.11,6.96-6.96V74.82C195.21,72.97,194.47,71.19,193.17,69.88L193.17,69.88z\n M179.15,78.02h-40.96V37.06L179.15,78.02z M179.54,200.33H57.67V36.67h65.73v47.01c0,5.05,4.09,9.14,9.14,9.14h47.01V200.33z\n M119.06,133.32l-13.45-22.29c-0.48-0.78-2.24-1.24-2.24-1.24h-8.33c-0.5,0-0.98,0.13-1.39,0.41c-1.21,0.76-1.58,2.36-0.8,3.6\n l17.85,28.28l-18.08,28.8c-0.76,1.22-0.39,2.83,0.84,3.6c0.41,0.26,0.89,0.39,1.37,0.39h7.48c0,0,1.74-0.26,2.22-1.02l13.65-22.09\n l13.56,22.07c0.48,0.78,2.22,1.26,2.22,1.26h8.18c0.5,0,0.98-0.15,1.42-0.42c1.22-0.79,1.57-2.4,0.79-3.63l-18.35-28.48\n l18.63-28.94c0.78-1.23,0.42-2.85-0.81-3.63c-0.42-0.27-0.9-0.41-1.4-0.41h-7.79c0,0-1.76,0.7-2.24,1.48L119.06,133.32z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\"\n >Full Nucleosome Panel List</span\n >\n </a>\n </div>\n </div>\n </div>\n </div>\n </div>\n</div>\n","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$1,795.00","value":1795,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"dCypher Nucleosome Full Panel for Enzyme Screening and HTS Assays","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"shipping_messages":[],"rating":0,"meta_keywords":"dcypher, dycpher panel, nuclesome panel, nucleosome array, 16-9001","show_quantity_input":1,"title":"dCypher™ Nucleosome Full Panel","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/662/975/dcypher-nucleosome-full-panel__10296.1645734567.jpg?c=2","alt":"dCypher Nucleosome Full Panel"}]}
Description
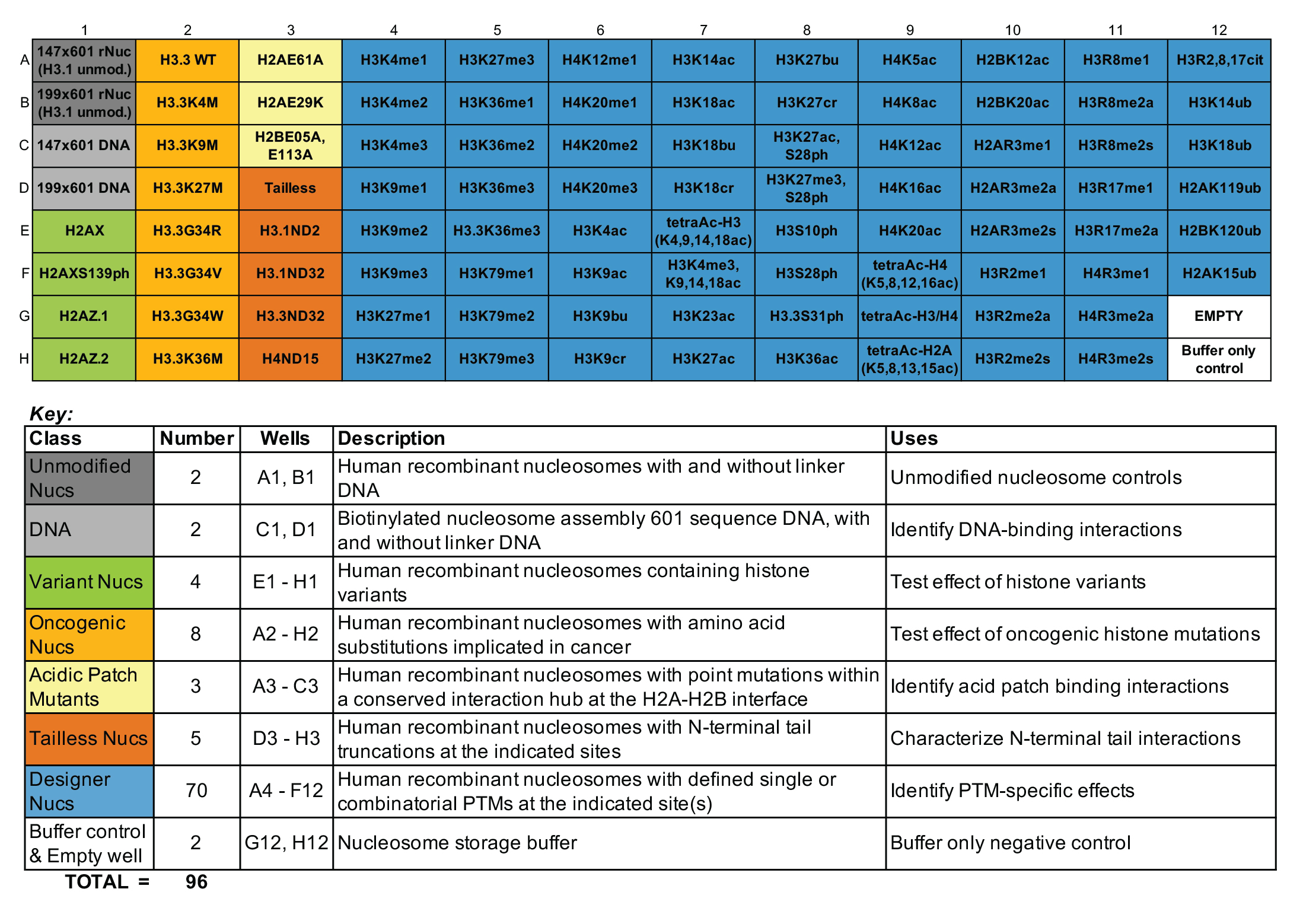
Understanding epigenetic reader proteins and their binding preference for post-translational modifications (PTMs) is key to unveiling how these proteins regulate genome processes and how their dysregulation may be contributing to disease. The dCypher™ Nucleosome Panel enables access to epigenetic diversity in a physiologically relevant context, the nucleosome, where chromatin reader activity can be accurately interrogated. The 96-well plate contains recombinant mononucleosomes as well as appropriate controls. Human histones expressed in E. coli bearing single and combinatorial histone post-translational modifications (PTMs) are wrapped by 147 or 199 base pair DNA with a 5’ biotin-TEG group and a central 601-positioning sequence, identified by Lowary and Widom [1].
All nucleosomes in the panel are subjected to EpiCypher’s rigorous quality control metrics, including: ESI-TOF mass spectrometry analysis of the modified histones, SDS-PAGE to confirm octamer composition and purity, native PAGE to confirm nucleosome assembly, and western blot analysis of the PTM, histone mutation, or histone variant (if applicable). For the full list of nucleosomes in the panel, including individual catalog numbers of full-size (50 µg) products, see the Documents & Resources section for the associated Excel sheet.
Validation Data
Figure 1: dCypher nucleosome full panel plate layout and
key
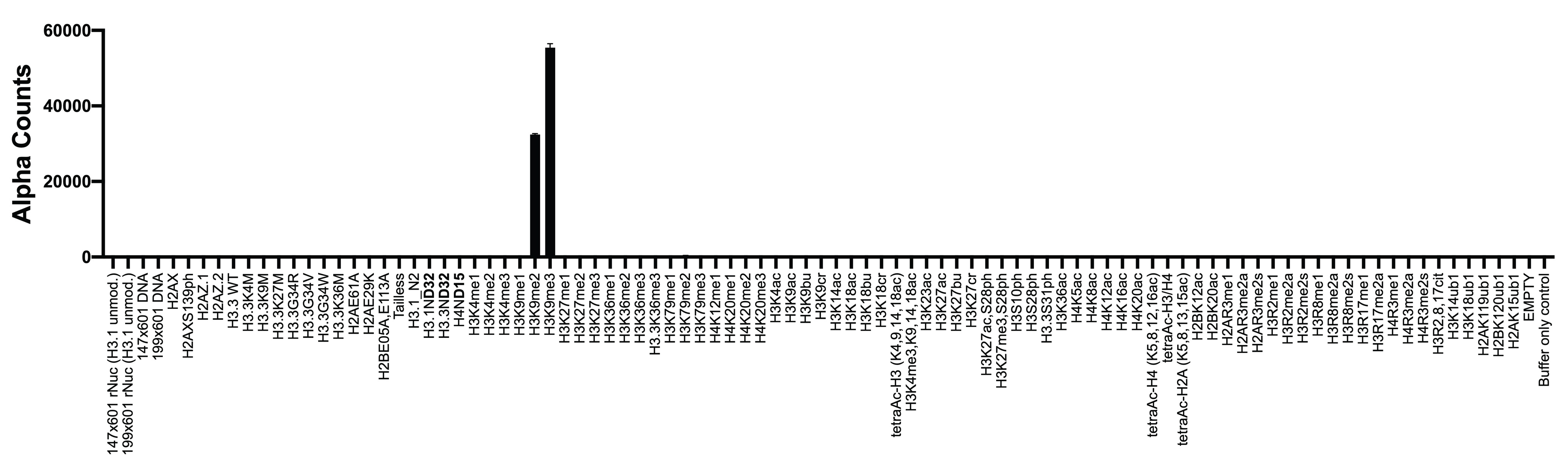
Figure 2: Chromatin reader binding data
GST-tagged HP1β (100 nM, EpiCypher
15-0074) was assayed with AlphaScreen (PerkinElmer) technology to
measure binding to nucleosomes (x-axis) in the dCypher
nucleosome Panel (10 nM). HP1β shows binding to the H3K9me3
and H3K9me2 dNucs, consistent with its reported binding
preference [5].
Technical Information
Gene & Protein Information
H2B - O60814 (alt. names: H2B K, HIRA-interacting protein 1)
H3.1 - P68431 (alt. names: H3, H3/a, H3/b, H3/c, H3/d)
H3.2 - Q71DI3
H3.3 - P84243
H4 - P62805
Application Notes
Access to epigenetic diversity in the context of a physiological nucleosome enables broad end-user applications, including nucleosome binding studies (e.g. chromatin reader binding preferences [2-4] see Figure 2), enzyme screening assays (e.g. identification of preferred substrates), and antibody specificity testing (e.g. for applications where histone peptide specificity is an insufficient surrogate). The biotin group on the DNA facilitates a wide variety of applications involving streptavidin capture.
Frequently Asked Questions
Q:
How many assay points of each nucleosome are supplied?
A: This varies according to the sensitivity of the
end application. For some context, the supplied format is sufficient
to perform >35 AlphaScreen assays, >30 IP-qPCR experiments, and 3-5
immunoblotting experiments.
Q:
How do I store the plate? Can I dilute the nucleosomes?
A: The nucleosomes are stable for 6 months at -80ºC
in the supplied format (1.5 µM). Freeze/thaw cycles should be
limited (2-3 times). When diluted below 1.5 µM, a carrier protein
(e.g. BSA) must be used to supplement the concentration, otherwise,
stability is affected. For example, in our SNAP-ChIP product line,
nucleosomes are diluted to 0.6 nM in 10 mM sodium cacodylate pH 7.5,
100 mM NaCl, 1 mM EDTA, 50% glycerol (w/v), 1x Protease Inhibitor
cocktail, 100 μg/mL BSA, 10 mM β-mercaptoethanol. In this
formulation, the nucleosomes are stable at -20ºC (where they do not
freeze) for 6 months.
For applications that may not be compatible with the SNAP-ChIP buffer, such as AlphaScreen, EpiCypher routinely dilutes nucleosomes to 80 nM in assay buffer (20 mM Tris pH 7.5, 0.01% BSA, 0.01% NP-40, 2mM DTT). Under these conditions the nucleosomes are stable for ~2 weeks at -80°C.
*Note: extremely high amounts of salt (>650 mM) and ionic detergents (e.g. SDS) will disrupt nucleosome stability.
References
[1] Lowary & Widom J. Mol. Biol. (1998). PMID: 9514715
[2] Weinberg et al. Nature (2019). PMID: 31485078
[3] Weinberg et al. Nat. Genet. (2021). PMID: 33986537
[4] Dilworth et al. Nat. Chem. Biol (2022). PMID: 34782742
[5] Albanese et al. ACS Chem. Biol. (2020). PMID: 31634430
Product References: