H3K27ac Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag
{"url":"https://www.epicypher.com/products/antibodies/h3k27ac-antibody-snap-certified-for-cut-run-and-cut-tag","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"id":1135,"bulk_discount_rates":[],"can_purchase":true,"meta_description":"Rabbit monoclonal histone H3K27ac antibody rigorously tested for robust and reliable performance in CUT&RUN and CUT&Tag","category":["Antibodies","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays","Antibodies/CUTANA™ CUT&RUN Antibodies","Antibodies/CUTANA™ CUT&RUN Antibodies/CUTANA™ CUT&RUN Antibodies to Histone PTMs","Antibodies/CUTANA™ CUT&Tag Antibodies"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1135/1203/snap-chip-ab__70507.1557259520.1280.1280__90586.1575483123.1280.1280__10437.1702325040.png?c=2","alt":"H3K27ac Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1135","shipping":{"calculated":true},"num_reviews":0,"weight":"0.01 LBS","custom_fields":[{"id":"1293","name":"Pack Size","value":"100 µg"}],"sku":"13-0059","description":"<div class=\"product-general-info\">\n <ul class=\"product-general-info__list-left\">\n <li class=\"product-general-info__list-item\">\n <strong>Type: </strong>Monoclonal [2114-3E4]\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Host: </strong>Rabbit\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Applications: </strong>CUT&RUN, CUT&Tag\n </li>\n </ul>\n <ul class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <strong>Reactivity: </strong>Human, Wide Range\n (Predicted)\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Format: </strong>Protein A affinity-purified\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Target Size: </strong>15 kDa\n </li>\n </ul>\n</div>\n\n<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <p>\n This H3K27ac (histone H3 acetylated at lysine 27) antibody meets EpiCypher’s lot-specific SNAP-Certified™\n criteria for specificity and efficient target enrichment in both CUT&RUN and CUT&Tag applications. This\n requires <20% cross-reactivity to related histone PTMs determined using the SNAP-CUTANA™ K-AcylStat Panel of\n spike-in controls (EpiCypher RD193002, <strong>Figures 1 and 5</strong>). High target efficiency is\n confirmed by consistent\n genomic enrichment at varying cell inputs: 500k and 50k cells in CUT&RUN (<strong>Figures 2-3</strong>);\n 100k and 10k cells\n in CUT&Tag (<strong>Figures 6-7</strong>). High efficiency antibodies display similar peak structures\n (<strong>Figures 3 and 7</strong>) and\n highly conserved genome-wide signal (<strong>Figures 2 and 6</strong>) even at reduced cell numbers.\n H3K27ac is associated\n with gene activation and is enriched at active enhancers and promoters [1]. </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data - CUT&RUN</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/antibodies/13-0059_snap_specificity_cr.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"13-0059_snap_specificity_cr\"\n src=\"/content/images/products/antibodies/13-0059_snap_specificity_cr.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 1: Average SNAP specificity analysis from two\n CUT&RUN experiments</strong><br />\n CUT&RUN was performed as described in <strong>Figure 4</strong>. CUT&RUN sequencing reads were\n aligned to the unique DNA\n barcodes corresponding to each nucleosome in the K-AcylStat panel (x-axis). Data are expressed as a\n percent relative to on-target recovery (H3K27ac set to 100%). The antibody showed recovery of\n H3K27ac spike-in nucleosomes as well as H3K27ac nucleosomes that contain a proximal phosphorylation\n at S28 at both 500k and 50k cells. The antibody cross-reacts with extended acyl states (butyrylation\n and crotonylation) at H3K27, but these are typically low abundance in cells [2].\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/antibodies/13-0059_genome-wide_enrichment_cr.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"13-0059_genome-wide_enrichment_cr\"\n src=\"/content/images/products/antibodies/13-0059_genome-wide_enrichment_cr.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 2: CUT&RUN genome-wide\n enrichment</strong><br />\n CUT&RUN was performed as described in\n <strong>Figure 4</strong>. Sequence reads were aligned\n to 18,793 annotated transcription start sites (TSSs ±\n 2 kbp). Signal enrichment was sorted from highest to\n lowest (top to bottom) relative to the H3K27ac - 500k\n cells reaction (all gene rows aligned). High, medium,\n and low intensity are shown in red, yellow, and blue,\n respectively. H3K4me3 positive control and H3K27ac\n antibodies produced the expected enrichment pattern,\n which was consistent between 500k and 50k cells and\n greater than the IgG negative control.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/antibodies/13-0059_browser_tracks_cr.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\">\n <img loading=\"lazy\" alt=\"13-0059_browser_tracks_cr\"\n src=\"/content/images/products/antibodies/13-0059_browser_tracks_cr.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span>\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 3: H3K27ac CUT&RUN representative\n browser tracks</strong><br />\n CUT&RUN was performed as described in\n <strong>Figure 4</strong>. Gene browser shots were\n generated using the Integrative Genomics Viewer (IGV,\n Broad Institute). H3K27ac antibody tracks display peaks at promoters and enhancers, consistent with\n the biological function of this PTM. Similar results in peak structure and\n location were observed for both 500k and 50k cell\n inputs.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0059-figure4.png?t=1700554861\"\n target=\"_blank\" class=\"image-picker__main-image-link\">\n <img loading=\"lazy\" alt=\"cut-and-run-methods\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0059-figure4.png?t=1700554861\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span>\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 4: CUT&RUN methods</strong><br />\n CUT&RUN was performed on 500k and 50k K562 cells with\n the SNAP-CUTANA™ K-MetStat Panel (EpiCypher\n <a href=\"/products/nucleosomes/snap-cutana-k-metstat-panel\" target=\"_blank\">19-1002</a>) or\n SNAP-CUTANA™ K-AcylStat Panel (EpiCypher\n RD193002) spiked-in prior to the addition of 0.5 µg of\n either IgG negative control (EpiCypher\n <a href=\"/products/nucleosomes/snap-cutana-spike-in-controls/cutana-rabbit-igg-cut-run-negative-control-antibody\"\n target=\"_blank\">13-0042</a>), H3K4me3 positive control (EpiCypher\n <a href=\"/products/antibodies/snap-chip-certified-antibodies/histone-h3k4me3-antibody-snap-chip-certified-cutana-cut-run-compatible\"\n target=\"_blank\">13-0041</a>), or H3K27ac antibodies. The experiment was\n performed using the CUTANA™ ChIC/CUT&RUN Kit v3\n (EpiCypher\n <a href=\"/products/epigenetics-reagents-and-assays/cutana-chic-cut-and-run-kit\"\n target=\"_blank\">14-1048</a>). Library preparation was performed with 5 ng of\n CUT&RUN enriched DNA (or the total amount recovered if\n less than 5 ng) using the CUTANA™ CUT&RUN Library Prep\n Kit (EpiCypher\n <a href=\"/products/epigenetics-reagents-and-assays/cutana-cut-and-run-library-prep-kit\"\n target=\"_blank\">14-1001/14-1002</a>). Both kit protocols were adapted for high\n throughput Tecan liquid handling. Libraries were run\n on an Illumina NextSeq2000 with paired-end sequencing\n (2x50 bp). Sample sequencing depth was 3.8 million reads (IgG 500k cell input), 5.0 million reads\n (IgG 50k cell input), 2.5 million reads (H3K4me3 500k cell input), 9.1 million reads (H3K4me3 50k\n cell input), 12.6 million reads (H3K27ac 500k cell input), and 11.0 million reads (H3K27ac 50k cell\n input). Data were aligned to the hg19 genome\n using Bowtie2. Data were filtered to remove\n duplicates, multi-aligned reads, and ENCODE DAC\n Exclusion List regions.\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img loading=\"lazy\" alt=\"13-0059_snap_specificity_cr\"\n src=\"/content/images/products/antibodies/13-0059_snap_specificity_cr.jpeg\" width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\" role=\"button\" />\n <img loading=\"lazy\" alt=\"13-0059-genome-wide-enrichment\"\n src=\"/content/images/products/antibodies/13-0059_genome-wide_enrichment_cr.jpeg\"\n class=\"image-picker__side-image\" role=\"button\" />\n <img loading=\"lazy\" alt=\"13-0059-representative-browser-tracks\"\n src=\"/content/images/products/antibodies/13-0059_browser_tracks_cr.jpeg\"\n class=\"image-picker__side-image\" role=\"button\" />\n <img loading=\"lazy\" alt=\"cut-and-run-methods\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0059-figure4.png?t=1700554861\"\n class=\"image-picker__side-image\" role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data - CUT&Tag</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <p></p>\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/antibodies/13-0059_snap_specificity_ct.jpeg\" target=\"_blank\"\n class=\"image-picker__main-image-link\">\n <img loading=\"lazy\" alt=\"13-0059_snap_specificity_ct\"\n src=\"/content/images/products/antibodies/13-0059_snap_specificity_ct.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span>\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 5: SNAP specificity analysis in CUT&Tag</strong><br />\n CUT&Tag was performed as described in <strong>Figure 8</strong>. CUT&Tag sequencing reads were\n aligned to the unique DNA\n barcodes corresponding to each nucleosome in the K-AcylStat panel (x-axis). Data are expressed as a\n percent relative to on-target recovery (H3K27ac set to 100%). The antibody showed recovery of\n H3K27ac spike-in nucleosomes and to a lesser extent H3K27ac nucleosomes that contain a proximal\n phosphorylation at S28 at both 500k and 50k cells. The antibody cross-reacts with butyrylation at\n H3K27, but this is typically low abundance in cells [2].\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/antibodies/13-0059_genome-wide_enrichment_ct.jpeg\"\n target=\"_blank\" class=\"image-picker__main-image-link\">\n <img loading=\"lazy\" alt=\"13-0059_genome-wide_enrichment_cr\"\n src=\"/content/images/products/antibodies/13-0059_genome-wide_enrichment_ct.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span>\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 6: CUT&Tag genome-wide enrichment</strong><br />\n CUT&Tag was performed as described in\n <strong>Figure 8</strong>. Sequence reads were aligned\n to 18,793 annotated transcription start sites (TSSs ±\n 2 kbp). Signal enrichment was sorted from highest to\n lowest (top to bottom) relative to the H3K27ac - 100k\n nuclei reaction (all gene rows aligned). High, medium,\n and low intensity are shown in red, yellow, and blue,\n respectively. H3K4me3 positive control and H3K27ac\n antibodies produced the expected enrichment pattern,\n which was consistent between 100k and 10k nuclei and\n greater than the IgG negative control.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/antibodies/13-0059_browser_tracks_ct.jpeg\"\n target=\"_blank\" class=\"image-picker__main-image-link\">\n <img loading=\"lazy\" alt=\"13-0059_browser_tracks_ct\"\n src=\"/content/images/products/antibodies/13-0059_browser_tracks_ct.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span>\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 7: H3K27ac CUT&Tag representative browser\n tracks</strong><br />\n CUT&Tag was performed as described in\n <strong>Figure 8</strong>. Gene browser shots were\n generated using the Integrative Genomics Viewer (IGV,\n Broad Institute). H3K27ac antibody tracks display peaks at promoters and enhancers, consistent with\n the biological function of this PTM. Similar results in peak structure and\n location were observed for both 100k and 10k nuclei\n inputs.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0059-figure8.png?t=1700554863\"\n target=\"_blank\" class=\"image-picker__main-image-link\">\n <img loading=\"lazy\" alt=\"cut-and-tag-methods\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0059-figure8.png?t=1700554863\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span>\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 8: CUT&Tag methods</strong><br />\n CUT&Tag was performed on 100k and 10k K562 nuclei with the SNAP-CUTANA™ K-MetStat Panel (EpiCypher\n <a href=\"/products/nucleosomes/snap-cutana-k-metstat-panel\" target=\"_blank\">19-1002</a>) or\n SNAP-CUTANA™ K-AcylStat Panel (EpiCypher RD193002) spiked-in prior to the addition of 0.5 µg of\n either IgG negative control (EpiCypher <a\n href=\"/products/nucleosomes/snap-cutana-spike-in-controls/cutana-rabbit-igg-cut-run-negative-control-antibody\"\n target=\"_blank\">13-0042</a>), H3K4me3 positive control (EpiCypher <a\n href=\"/products/antibodies/snap-chip-certified-antibodies/histone-h3k4me3-antibody-snap-chip-certified-cutana-cut-run-compatible\"\n target=\"_blank\">13-0041</a>), or H3K27ac antibodies. The experiment was performed using the\n CUTANA™ CUT&Tag Kit v1 (EpiCypher <a\n href=\"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-cut-and-tag-kit\"\n target=\"_blank\">14-1102/14-1103</a>). Libraries were run on an Illumina NextSeq2000 with\n paired-end sequencing (2x50 bp). Sample sequencing depth was 1.3 million reads (IgG 100k nuclei\n input), 2.0 million reads (IgG 10k nuclei input), 3.5 million reads (H3K4me3 100k nuclei input), 8.0\n million reads (H3K4me3 10k nuclei input), 8.1 million reads (H3K27ac 100k nuclei input) and 8.5\n million reads (H3K27ac 10k nuclei input). Data were aligned to the hg19 genome using Bowtie2. Data\n were filtered to remove\n duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img loading=\"lazy\" alt=\"13-0059_snap_specificity_ct\"\n src=\"/content/images/products/antibodies/13-0059_snap_specificity_ct.jpeg\"\n class=\"image-picker__side-image\" role=\"button\" />\n <img loading=\"lazy\" alt=\"13-0059_genome-wide_enrichment_ct\"\n src=\"/content/images/products/antibodies/13-0059_genome-wide_enrichment_ct.jpeg\"\n class=\"image-picker__side-image\" role=\"button\" />\n <img loading=\"lazy\" alt=\"13-0059_browser_tracks_ct\"\n src=\"/content/images/products/antibodies/13-0059_browser_tracks_ct.jpeg\"\n class=\"image-picker__side-image\" role=\"button\" />\n <img loading=\"lazy\" alt=\"cut-and-run-methods\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0059-figure8.png?t=1700554863\"\n class=\"image-picker__side-image\" role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Immunogen</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n A synthetic peptide corresponding to histone H3 acetylated\n at lysine 27\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Stable for 1 year at 4ºC from date of receipt\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Formulation</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Protein A affinity-purified recombinant monoclonal antibody in borate buffered\n saline pH 8.0, 0.09% sodium azide\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Recommended Dilution</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>CUT&RUN</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 0.5 µg per reaction\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>CUT&Tag</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 0.5 µg per reaction\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Gene & Protein Information</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Uniprot ID</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n H3.1 – P68431\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Alternate Names</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n H3, H3/a, H3/b, H3/d\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">References</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <strong>Background References:</strong>\n <br />\n [1] Pei et al. <i>Clinical Epigenetics</i> (2020). PMID:\n <a href=\"https://pubmed.ncbi.nlm.nih.gov/32664951/\" target=\"_blank\"\n title=\"H3K27ac acetylome signatures reveal the epigenomic reorganization in remodeled non-failing human hearts\">32664951</a>\n <br />\n [2] Simithy et al. <i>Nature Communications</i> (2017). PMID:\n <a href=\"https://pubmed.ncbi.nlm.nih.gov/29070843/\" target=\"_blank\"\n title=\"Characterization of histone acylations links chromatin modifications with metabolism\">29070843</a>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Documents & Resources</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-documents\">\n <a href=\"/content/documents/tds/13-0059.pdf\" target=\"_blank\" class=\"product-documents__link\">\n <svg version=\"1.1\" id=\"Layer_1\" xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\" x=\"0px\" y=\"0px\" viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\" xml:space=\"preserve\" class=\"product-documents__icon\"\n alt=\"16-0030 Datasheet\">\n <g>\n <path class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect x=\"20\" y=\"112\" class=\"product-documents__svg-background\" width=\"146\" height=\"76\" />\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Technical Datasheet</span>\n </a>\n </div>\n </div>\n </div>\n </div>\n </div>\n</div>","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$545.00","value":545,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"H3K27ac Antibody | SNAP-Certified for CUT&RUN and CUT&Tag","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"options":[],"related_products":[{"id":986,"sku":null,"name":"CUTANA™ CUT&Tag Kit","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-cut-and-tag-kit","availability":"","rating":null,"brand":{"name":null},"category":["Antibodies/CUTANA™ CUT&Tag Antibodies","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/986/1213/CTK20__46720.1713641110.png?c=2","alt":"CUTANA™ CUT&Tag Kit"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/986/1213/CTK20__46720.1713641110.png?c=2","alt":"CUTANA™ CUT&Tag Kit"}],"date_added":"8th Mar 2023","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1219,"name":"Pack Size","value":"48 Reactions"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$2,695.00","value":2695},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=986"},{"id":694,"sku":null,"name":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows","url":"https://www.epicypher.com/products/epigenetics-reagents-and-assays/cutana-pag-mnase-for-chic-cut-and-run-workflows","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n Type: Nuclease\n \n \n Mol Wgt: 43.7 kDa\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/694/689/Screen_Shot_2020-02-12_at_11.01.55_AM__17144.1581530752.png?c=2","alt":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/694/689/Screen_Shot_2020-02-12_at_11.01.55_AM__17144.1581530752.png?c=2","alt":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows"}],"date_added":"12th Aug 2019","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1174,"name":"Internal Comment","value":"Excess in bottom of Venom"},{"id":1175,"name":"Internal Comment","value":"Bulk in Psylocke"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$335.00","value":335},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$335.00","value":335},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$1,295.00","value":1295},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=694"},{"id":758,"sku":"13-0042","name":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag","url":"https://www.epicypher.com/products/nucleosomes/snap-cutana-spike-in-controls/cutana-igg-negative-control-antibody-for-cut-run-and-cut-tag","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes/SNAP-CUTANA™ Spike-in Controls","Antibodies/CUTANA™ CUT&RUN Antibodies","Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n Type: Polyclonal\n \n \n Host: Rabbit\n \n \n Applications: CUT&R","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/758/739/1579731877.1280.1280__61771.1592491196.png?c=2","alt":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/758/739/1579731877.1280.1280__61771.1592491196.png?c=2","alt":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag"}],"date_added":"17th Jun 2020","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1148,"name":"Pack Size","value":"100 µg"},{"id":1149,"name":"Internal Comment","value":"bulk stocks from Thermo in cold room"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=758","price":{"without_tax":{"currency":"USD","formatted":"$105.00","value":105},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=758"}],"shipping_messages":[],"rating":0,"meta_keywords":"H3K27ac antibody, histone H3K27ac antibody, histone H3 lysine 27 trimethylated antibody, CUT&RUN antibody, CUT&Tag antibody","show_quantity_input":1,"title":"H3K27ac Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1135/1203/snap-chip-ab__70507.1557259520.1280.1280__90586.1575483123.1280.1280__10437.1702325040.png?c=2","alt":"H3K27ac Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag"}]} Pack Size: 100 µg
- Type: Monoclonal [2114-3E4]
- Host: Rabbit
- Applications: CUT&RUN, CUT&Tag
- Reactivity: Human, Wide Range (Predicted)
- Format: Protein A affinity-purified
- Target Size: 15 kDa
Description
This H3K27ac (histone H3 acetylated at lysine 27) antibody meets EpiCypher’s lot-specific SNAP-Certified™ criteria for specificity and efficient target enrichment in both CUT&RUN and CUT&Tag applications. This requires <20% cross-reactivity to related histone PTMs determined using the SNAP-CUTANA™ K-AcylStat Panel of spike-in controls (EpiCypher RD193002, Figures 1 and 5). High target efficiency is confirmed by consistent genomic enrichment at varying cell inputs: 500k and 50k cells in CUT&RUN (Figures 2-3); 100k and 10k cells in CUT&Tag (Figures 6-7). High efficiency antibodies display similar peak structures (Figures 3 and 7) and highly conserved genome-wide signal (Figures 2 and 6) even at reduced cell numbers. H3K27ac is associated with gene activation and is enriched at active enhancers and promoters [1].
Validation Data - CUT&RUN
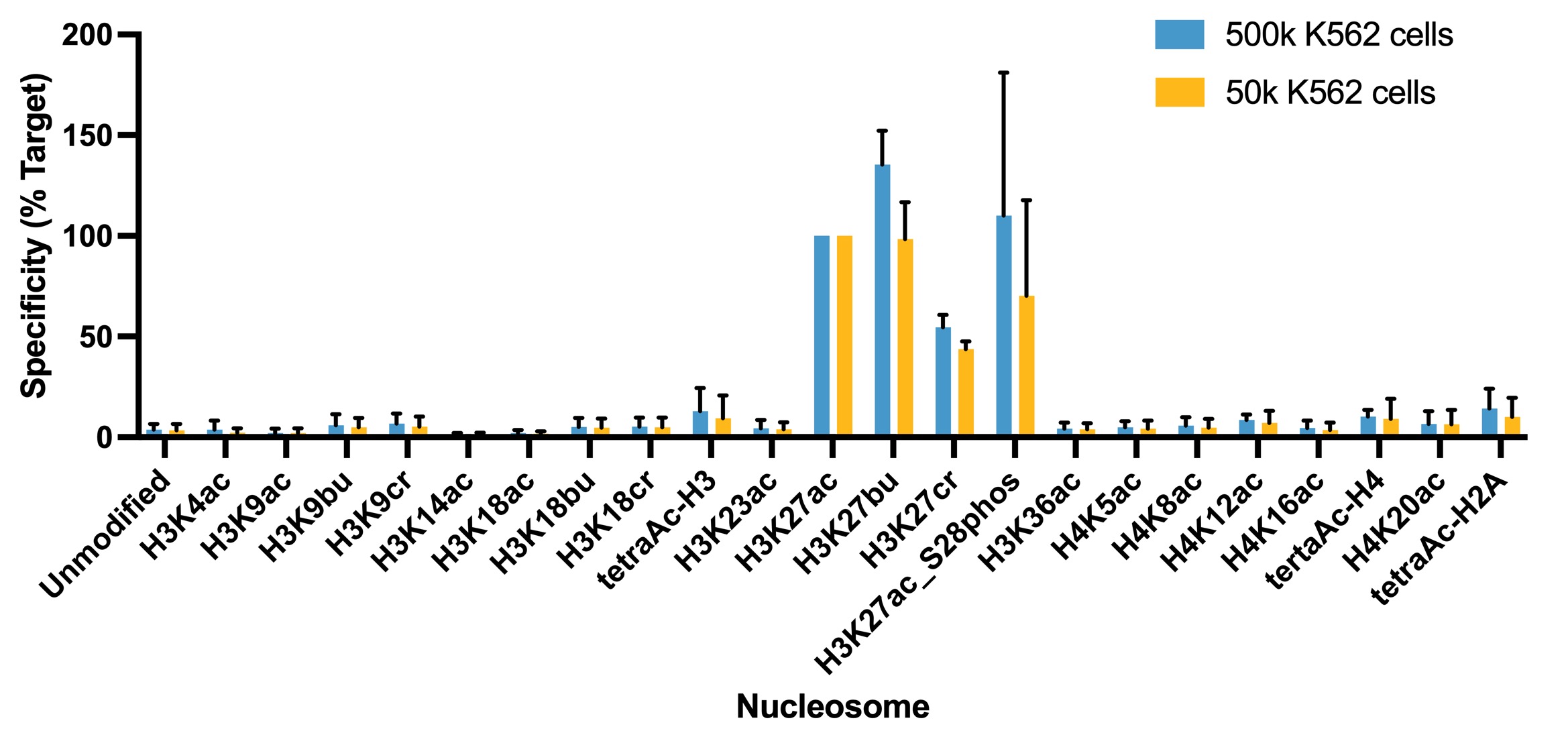
Figure 1: Average SNAP specificity analysis from two
CUT&RUN experiments
CUT&RUN was performed as described in Figure 4. CUT&RUN sequencing reads were
aligned to the unique DNA
barcodes corresponding to each nucleosome in the K-AcylStat panel (x-axis). Data are expressed as a
percent relative to on-target recovery (H3K27ac set to 100%). The antibody showed recovery of
H3K27ac spike-in nucleosomes as well as H3K27ac nucleosomes that contain a proximal phosphorylation
at S28 at both 500k and 50k cells. The antibody cross-reacts with extended acyl states (butyrylation
and crotonylation) at H3K27, but these are typically low abundance in cells [2].
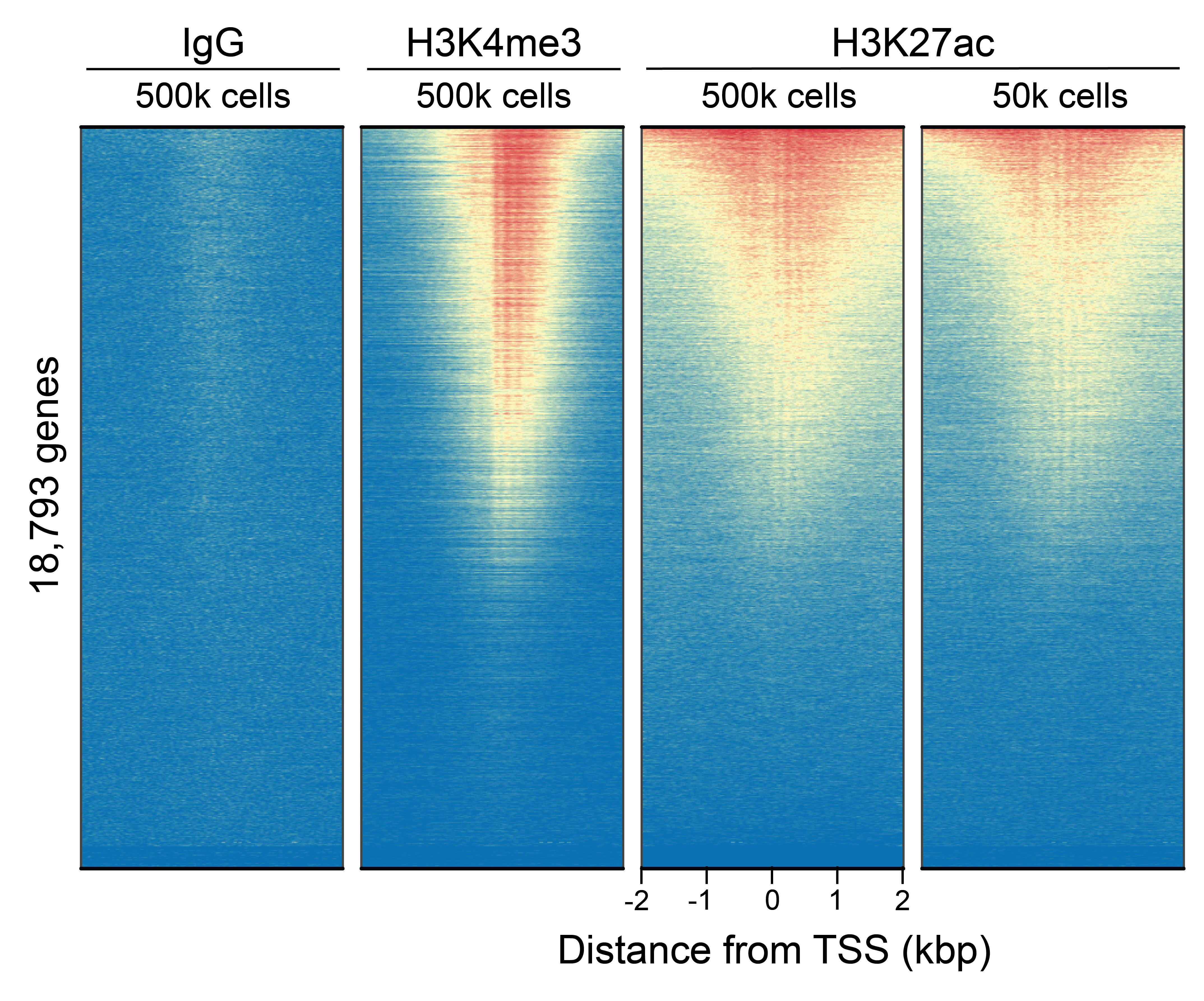
Figure 2: CUT&RUN genome-wide
enrichment
CUT&RUN was performed as described in
Figure 4. Sequence reads were aligned
to 18,793 annotated transcription start sites (TSSs ±
2 kbp). Signal enrichment was sorted from highest to
lowest (top to bottom) relative to the H3K27ac - 500k
cells reaction (all gene rows aligned). High, medium,
and low intensity are shown in red, yellow, and blue,
respectively. H3K4me3 positive control and H3K27ac
antibodies produced the expected enrichment pattern,
which was consistent between 500k and 50k cells and
greater than the IgG negative control.
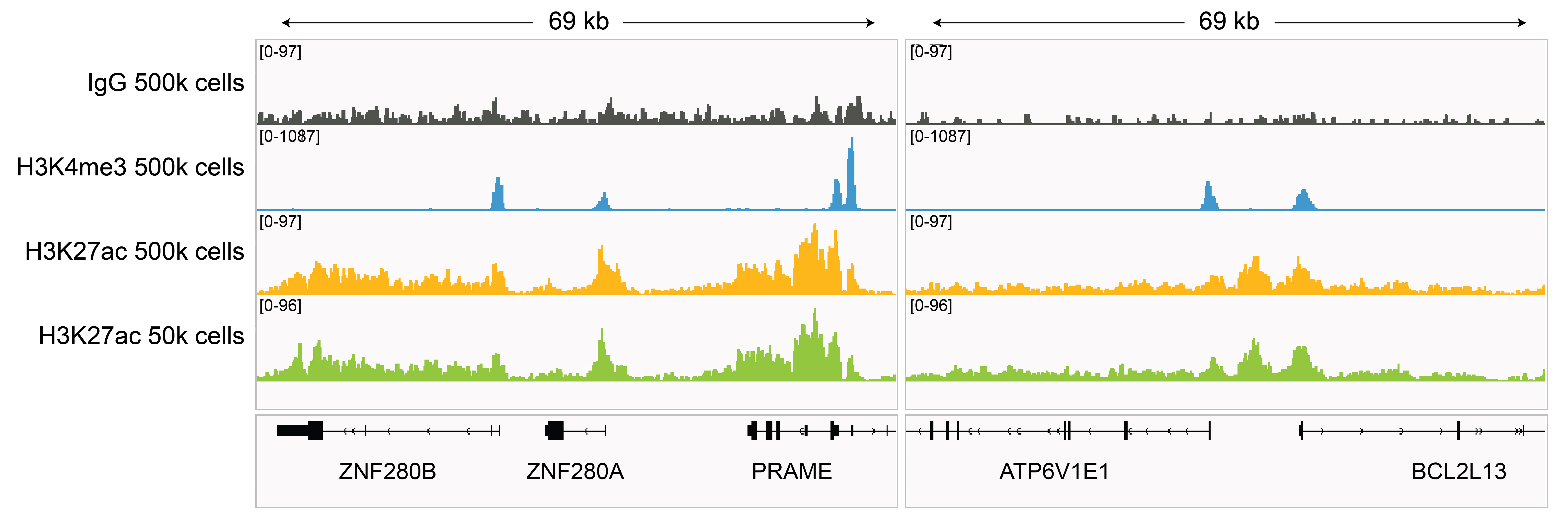
Figure 3: H3K27ac CUT&RUN representative
browser tracks
CUT&RUN was performed as described in
Figure 4. Gene browser shots were
generated using the Integrative Genomics Viewer (IGV,
Broad Institute). H3K27ac antibody tracks display peaks at promoters and enhancers, consistent with
the biological function of this PTM. Similar results in peak structure and
location were observed for both 500k and 50k cell
inputs.
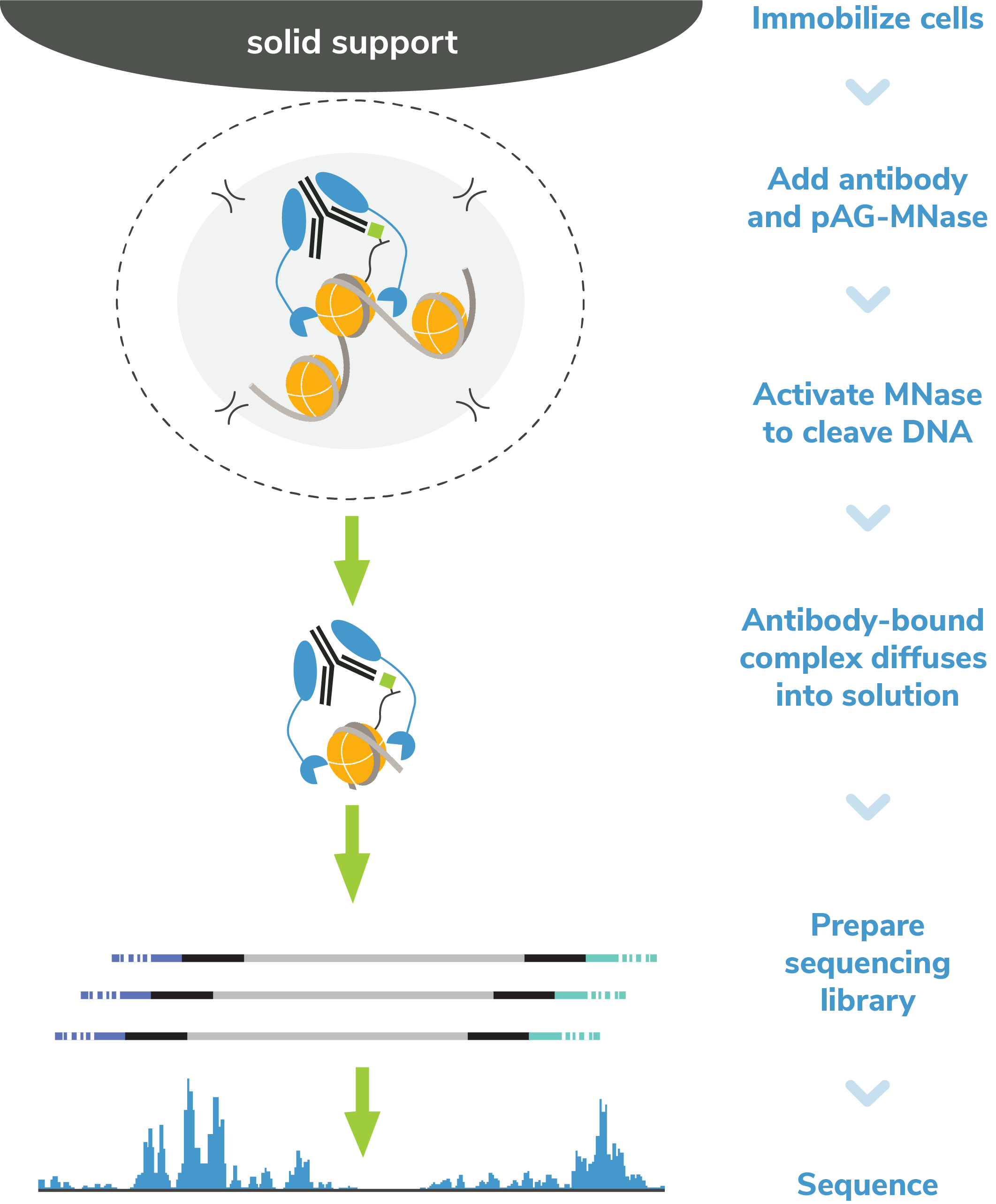
Figure 4: CUT&RUN methods
CUT&RUN was performed on 500k and 50k K562 cells with
the SNAP-CUTANA™ K-MetStat Panel (EpiCypher
19-1002) or
SNAP-CUTANA™ K-AcylStat Panel (EpiCypher
RD193002) spiked-in prior to the addition of 0.5 µg of
either IgG negative control (EpiCypher
13-0042), H3K4me3 positive control (EpiCypher
13-0041), or H3K27ac antibodies. The experiment was
performed using the CUTANA™ ChIC/CUT&RUN Kit v3
(EpiCypher
14-1048). Library preparation was performed with 5 ng of
CUT&RUN enriched DNA (or the total amount recovered if
less than 5 ng) using the CUTANA™ CUT&RUN Library Prep
Kit (EpiCypher
14-1001/14-1002). Both kit protocols were adapted for high
throughput Tecan liquid handling. Libraries were run
on an Illumina NextSeq2000 with paired-end sequencing
(2x50 bp). Sample sequencing depth was 3.8 million reads (IgG 500k cell input), 5.0 million reads
(IgG 50k cell input), 2.5 million reads (H3K4me3 500k cell input), 9.1 million reads (H3K4me3 50k
cell input), 12.6 million reads (H3K27ac 500k cell input), and 11.0 million reads (H3K27ac 50k cell
input). Data were aligned to the hg19 genome
using Bowtie2. Data were filtered to remove
duplicates, multi-aligned reads, and ENCODE DAC
Exclusion List regions.
Validation Data - CUT&Tag
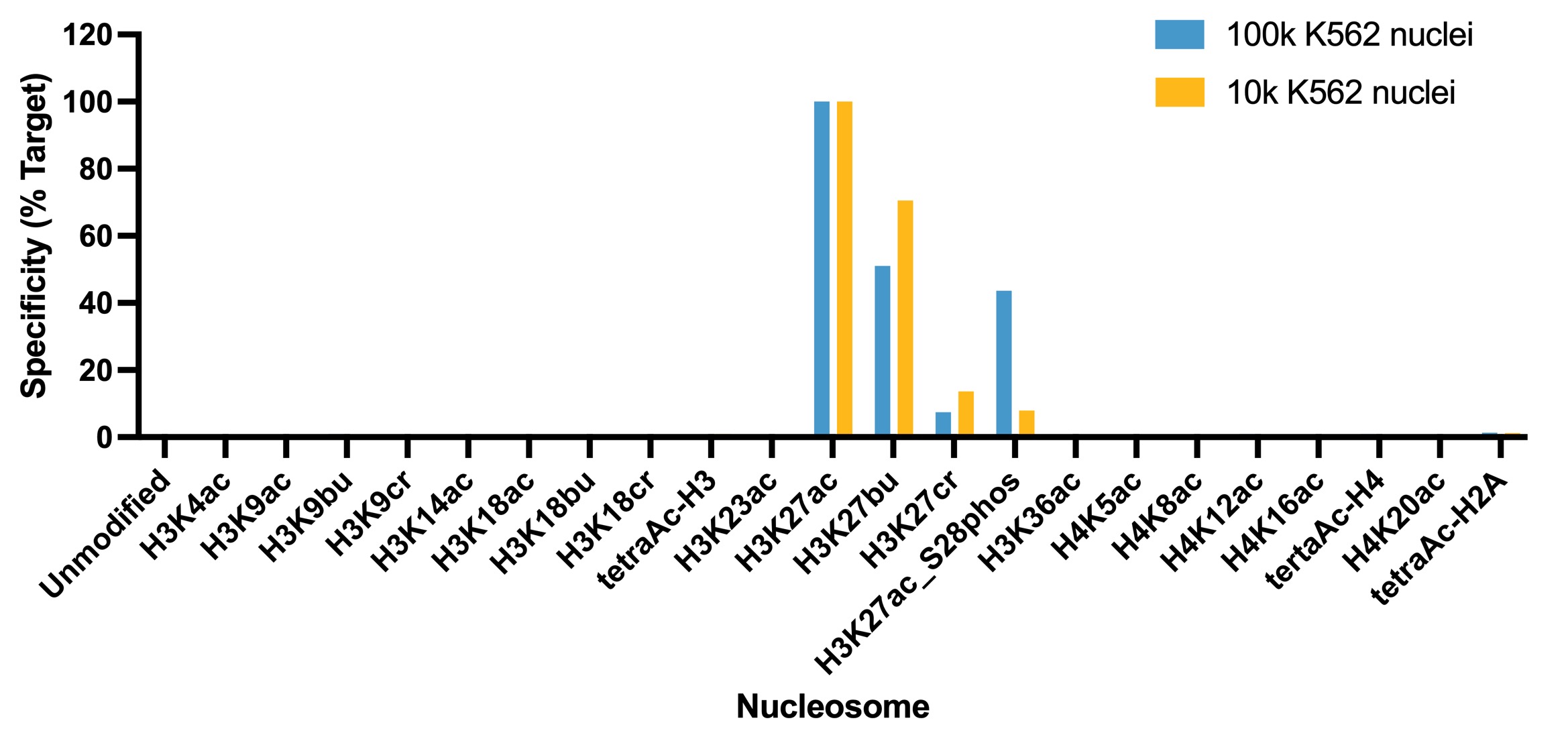
Figure 5: SNAP specificity analysis in CUT&Tag
CUT&Tag was performed as described in Figure 8. CUT&Tag sequencing reads were
aligned to the unique DNA
barcodes corresponding to each nucleosome in the K-AcylStat panel (x-axis). Data are expressed as a
percent relative to on-target recovery (H3K27ac set to 100%). The antibody showed recovery of
H3K27ac spike-in nucleosomes and to a lesser extent H3K27ac nucleosomes that contain a proximal
phosphorylation at S28 at both 500k and 50k cells. The antibody cross-reacts with butyrylation at
H3K27, but this is typically low abundance in cells [2].
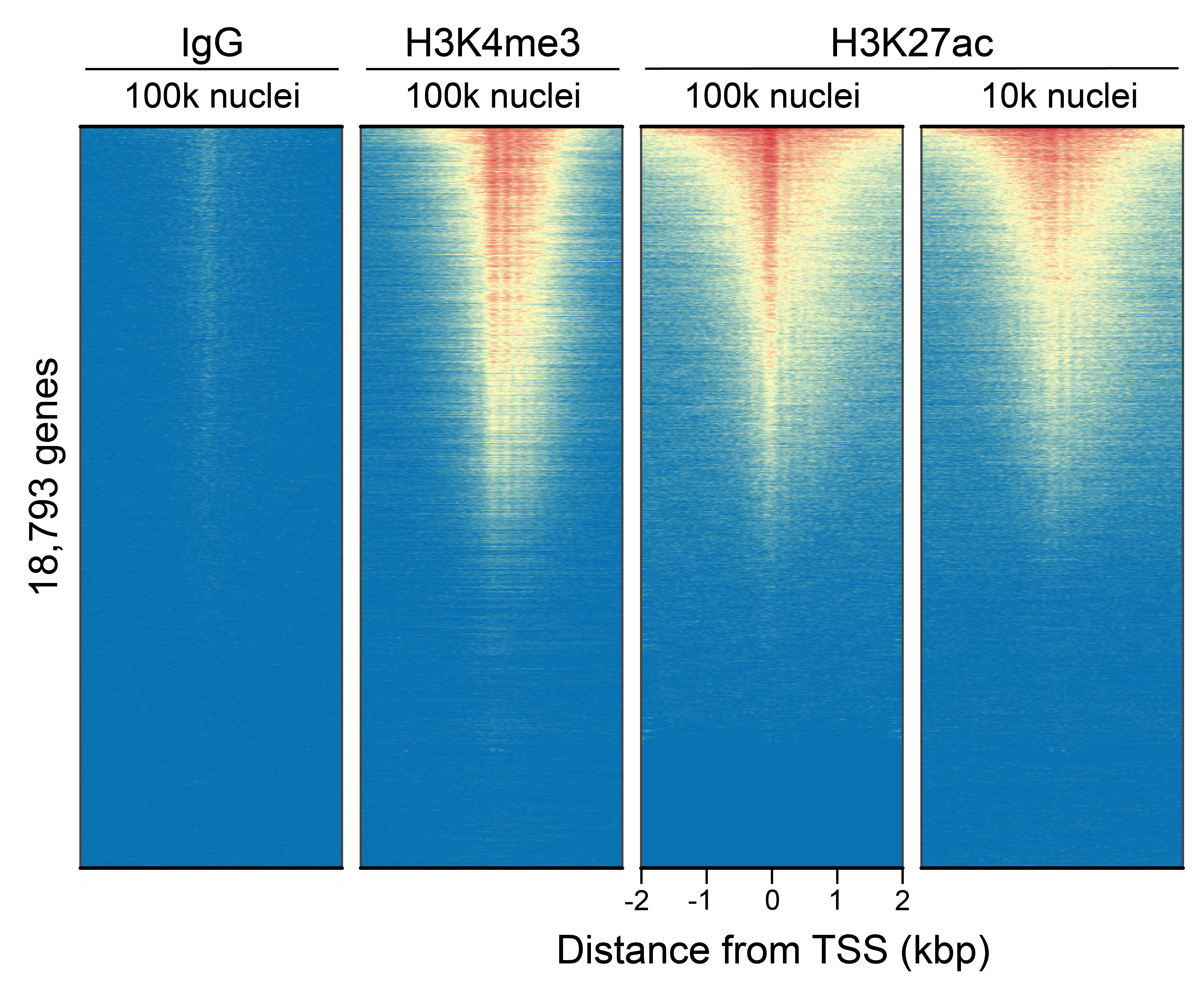
Figure 6: CUT&Tag genome-wide enrichment
CUT&Tag was performed as described in
Figure 8. Sequence reads were aligned
to 18,793 annotated transcription start sites (TSSs ±
2 kbp). Signal enrichment was sorted from highest to
lowest (top to bottom) relative to the H3K27ac - 100k
nuclei reaction (all gene rows aligned). High, medium,
and low intensity are shown in red, yellow, and blue,
respectively. H3K4me3 positive control and H3K27ac
antibodies produced the expected enrichment pattern,
which was consistent between 100k and 10k nuclei and
greater than the IgG negative control.
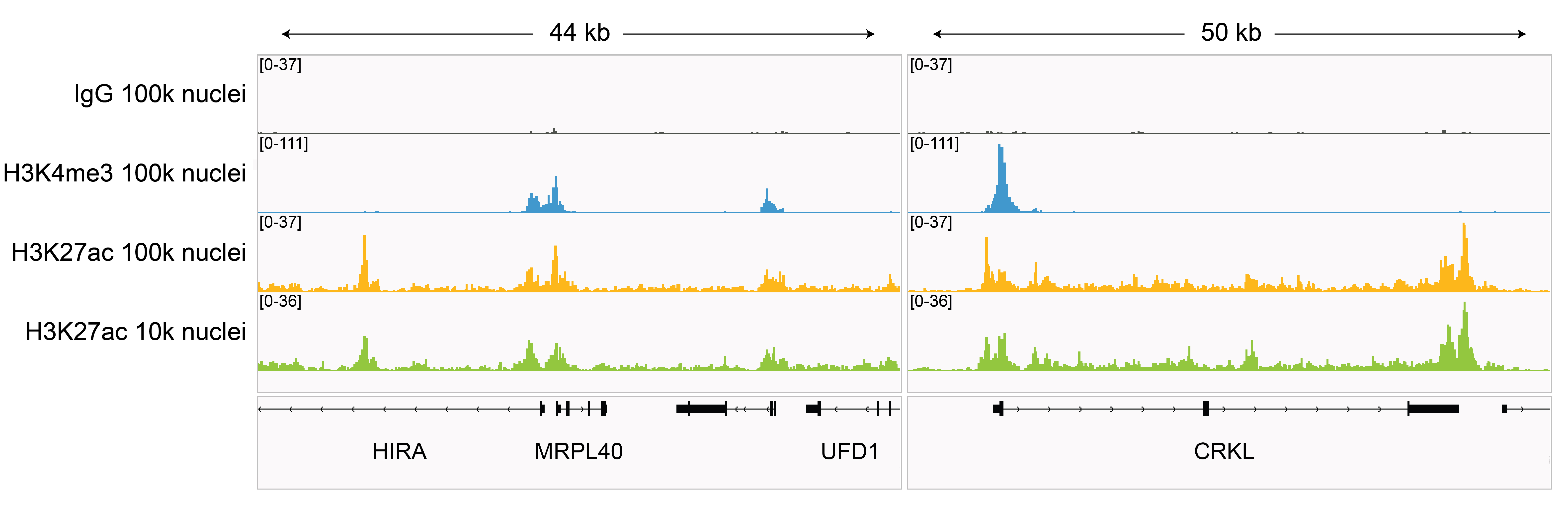
Figure 7: H3K27ac CUT&Tag representative browser
tracks
CUT&Tag was performed as described in
Figure 8. Gene browser shots were
generated using the Integrative Genomics Viewer (IGV,
Broad Institute). H3K27ac antibody tracks display peaks at promoters and enhancers, consistent with
the biological function of this PTM. Similar results in peak structure and
location were observed for both 100k and 10k nuclei
inputs.
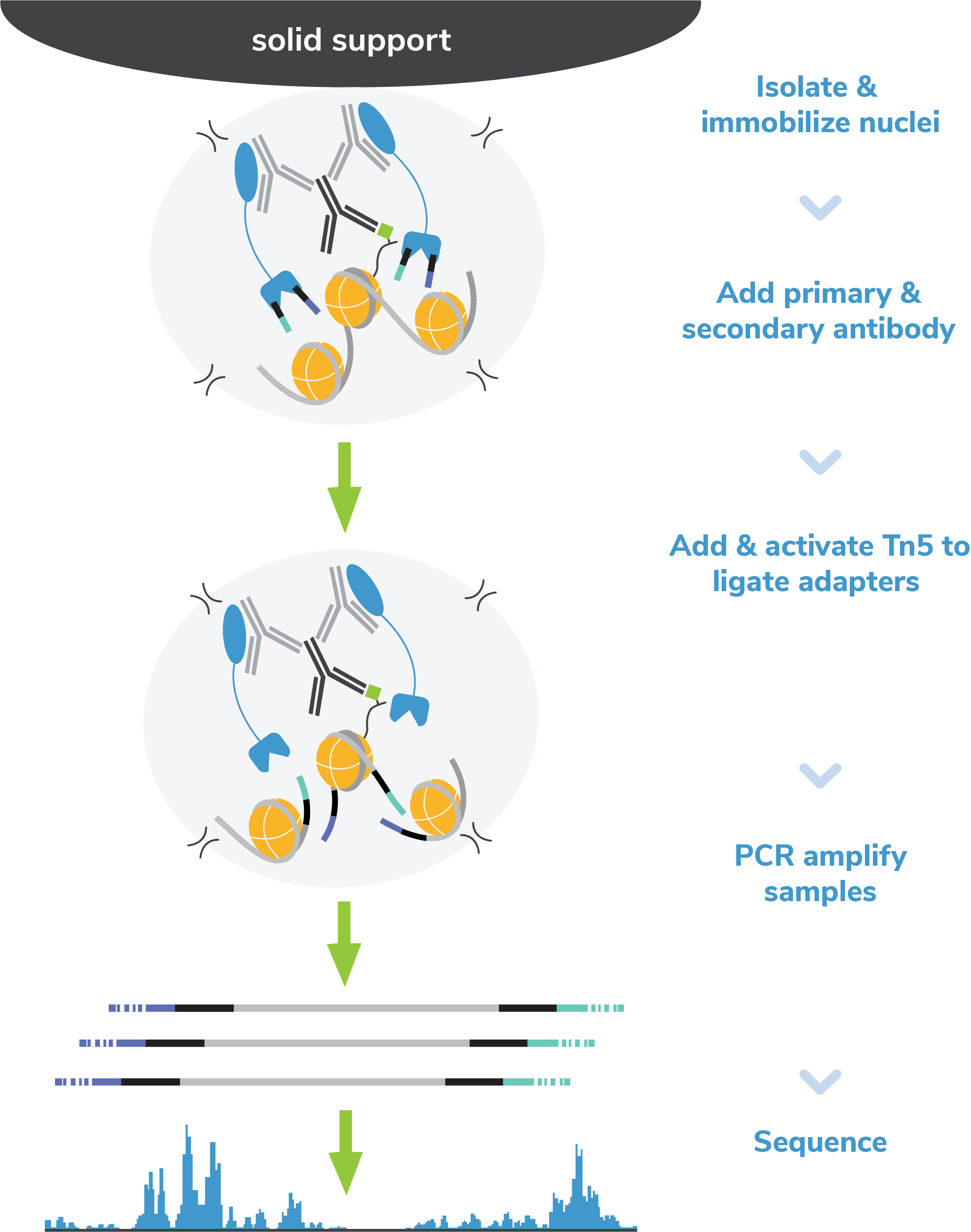
Figure 8: CUT&Tag methods
CUT&Tag was performed on 100k and 10k K562 nuclei with the SNAP-CUTANA™ K-MetStat Panel (EpiCypher
19-1002) or
SNAP-CUTANA™ K-AcylStat Panel (EpiCypher RD193002) spiked-in prior to the addition of 0.5 µg of
either IgG negative control (EpiCypher 13-0042), H3K4me3 positive control (EpiCypher 13-0041), or H3K27ac antibodies. The experiment was performed using the
CUTANA™ CUT&Tag Kit v1 (EpiCypher 14-1102/14-1103). Libraries were run on an Illumina NextSeq2000 with
paired-end sequencing (2x50 bp). Sample sequencing depth was 1.3 million reads (IgG 100k nuclei
input), 2.0 million reads (IgG 10k nuclei input), 3.5 million reads (H3K4me3 100k nuclei input), 8.0
million reads (H3K4me3 10k nuclei input), 8.1 million reads (H3K27ac 100k nuclei input) and 8.5
million reads (H3K27ac 10k nuclei input). Data were aligned to the hg19 genome using Bowtie2. Data
were filtered to remove
duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.