Extensively validated. Meticulously optimized.
Cleavage Under Targets and Release using Nuclease (CUT&RUN), based on the previous Chromatin ImmunoCleavage (ChIC) approach, uses a fusion of protein A, protein G, and micrococcal nuclease (pAG-MNase) to cleave and extract antibody-bound chromatin. The targeted nature of CUT&RUN's pAG-MNase improves signal : noise and reduces sequencing costs resulting in a highly cost- and time-efficient chromatin mapping assay.
EpiCypher's genomics experts have spent years developing and optimizing the CUT&RUN approach. We have built on the original protocol, adopting it for 8-strip tubes to maximize throughput, and developing robust spike-ins and quality control checks to ensure relaible results. The CUTANA™ CUT&RUN Kit is the culmination of these efforts, providing customers with a standout tool for accurate and user-friendly chromatin profiling.
To learn more about CUT&RUN, check out our Technical Support Center.
Have Questions?
We’re here to help. Click below and a member of our team will get back to you shortly!
Request More Info- Diverse target compatibility
- Low cell input requirements
- Highly robust and reproducible workflow
- Access to exclusive quality controls
- Multiple kit sizes to fit any project
Compatible with a wide variety of targets
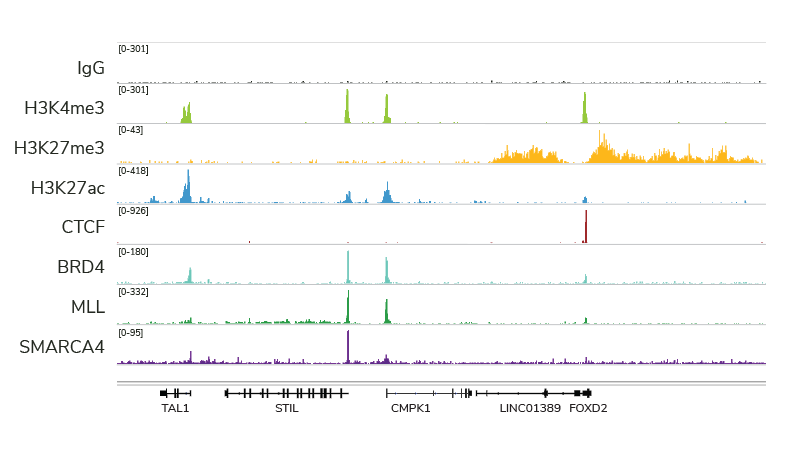
EpiCypher’s CUTANA ChIC/CUT&RUN Kit and associated protocol has been optimized for compatibility with diverse target proteins, including histone post-translational modifications, transcription factors, epigenetic enzymes and reader proteins, chromatin remodeling proteins. Each target produces a clear genomic enrichment pattern consistent with its known biological function, with little background.
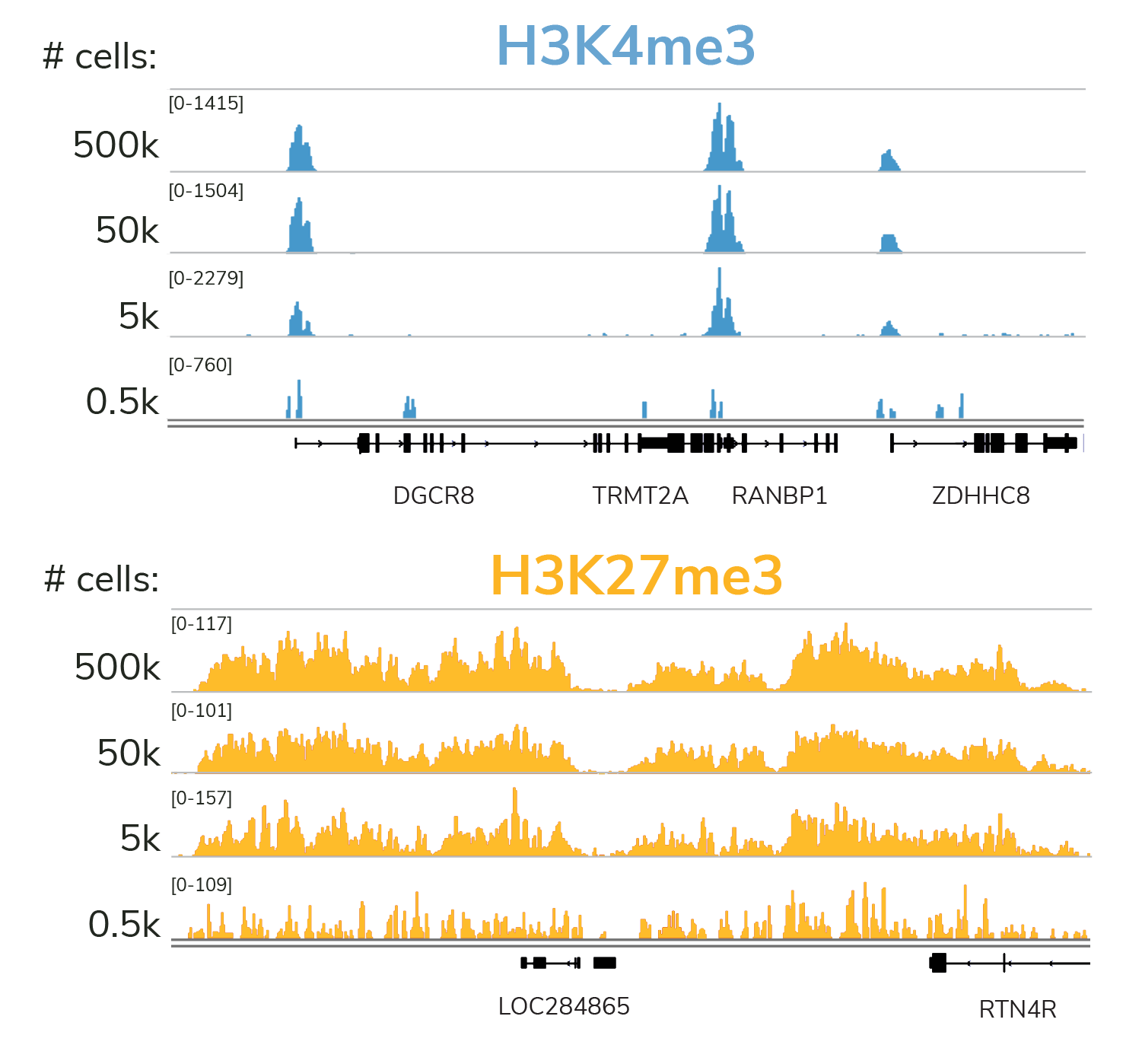
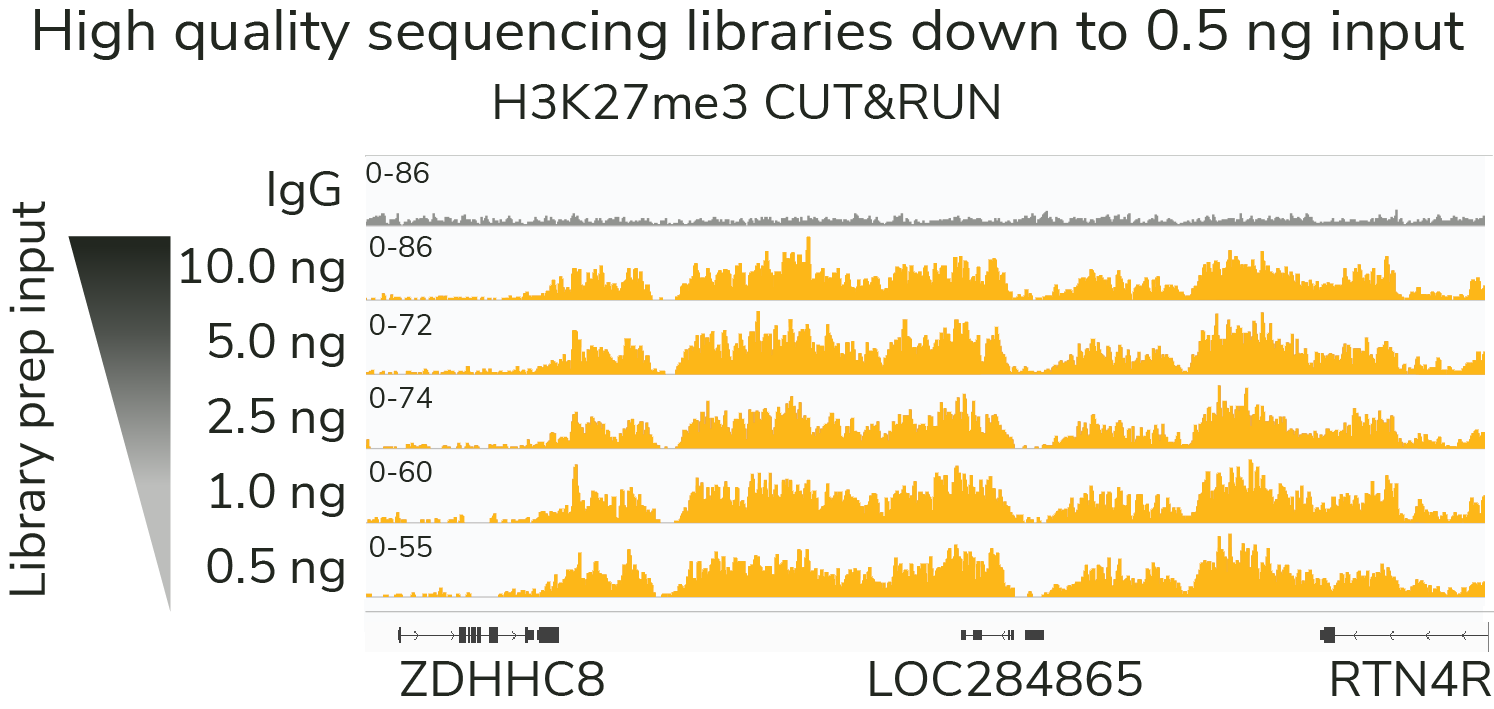
Quality data with low cell input
Our rigorously tested CUT&RUN protocol generates nearly identical data from 5,000 to 500,000 cells with no changes to the protocol. Furthermore, all of these data were generated using only 3 to 8 million reads per sample - a fraction of the sequencing depth required for ChIP-seq. Lower cost and sample input requirements allow for greater experimental throughput.
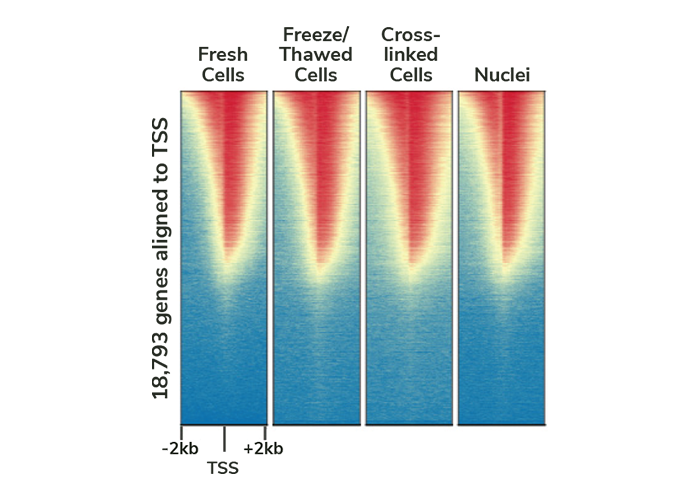
Validated sample preparation methods
We used our CUTANA ChIC/CUT&RUN Kit and adaptations as described in our manual to profile H3K4me3 in fresh, cryopreserved, and lightly cross-linked cells or nuclei. The resulting CUT&RUN data quality is indistinguishable across inputs, meaning you can apply CUT&RUN for more experiments and sample types.
Includes all the controls you need for a well-designed publication-quality experiment

E. coli Spike-in DNA
Monitor DNA loss and/or amplification during library preparation to normalize sequencing data and perform pairwise comparisons.
Control Antibodies
Lot-tested positive and negative control antibodies generate reliable CUT&RUN data and display strong correlation with ChIP-seq. Kit includes clear instructions for their application in CUT&RUN.

Spike-in Nucleosome Controls
First-in-class, defined DNA-barcoded nucleosome controls for CUT&RUN. Combine with positive and negative control antibodies to monitor and quantify experimental success.